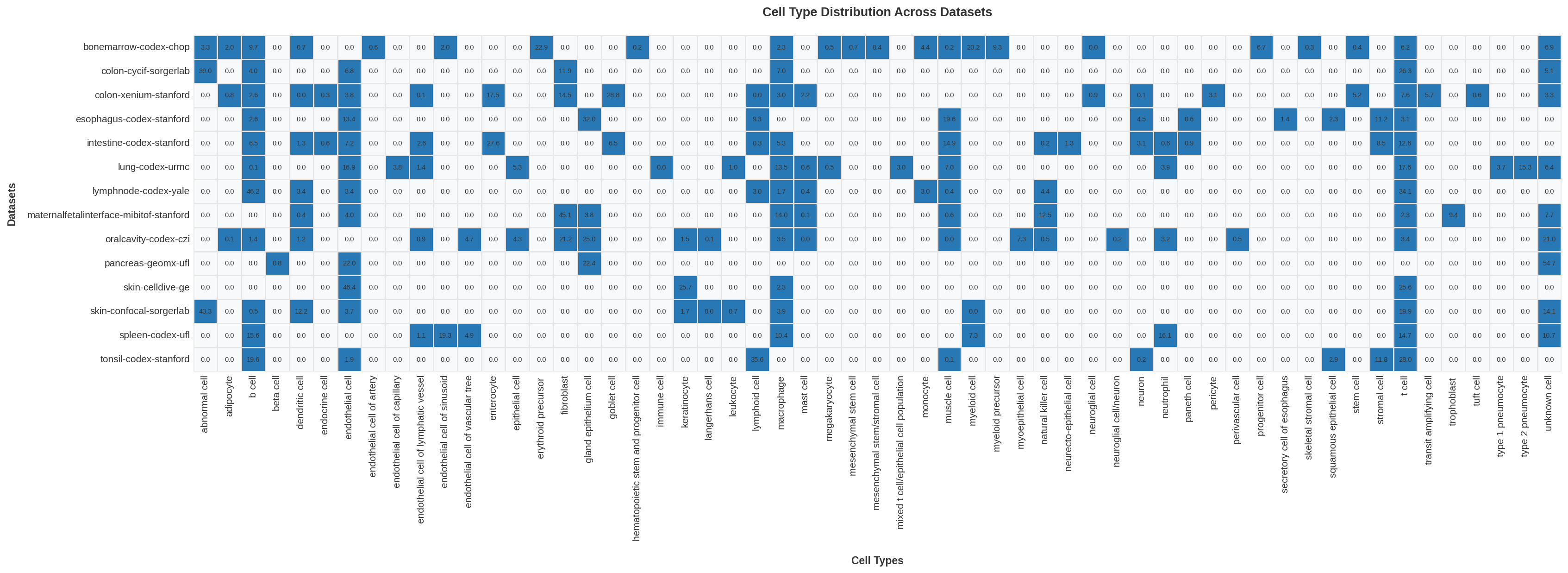
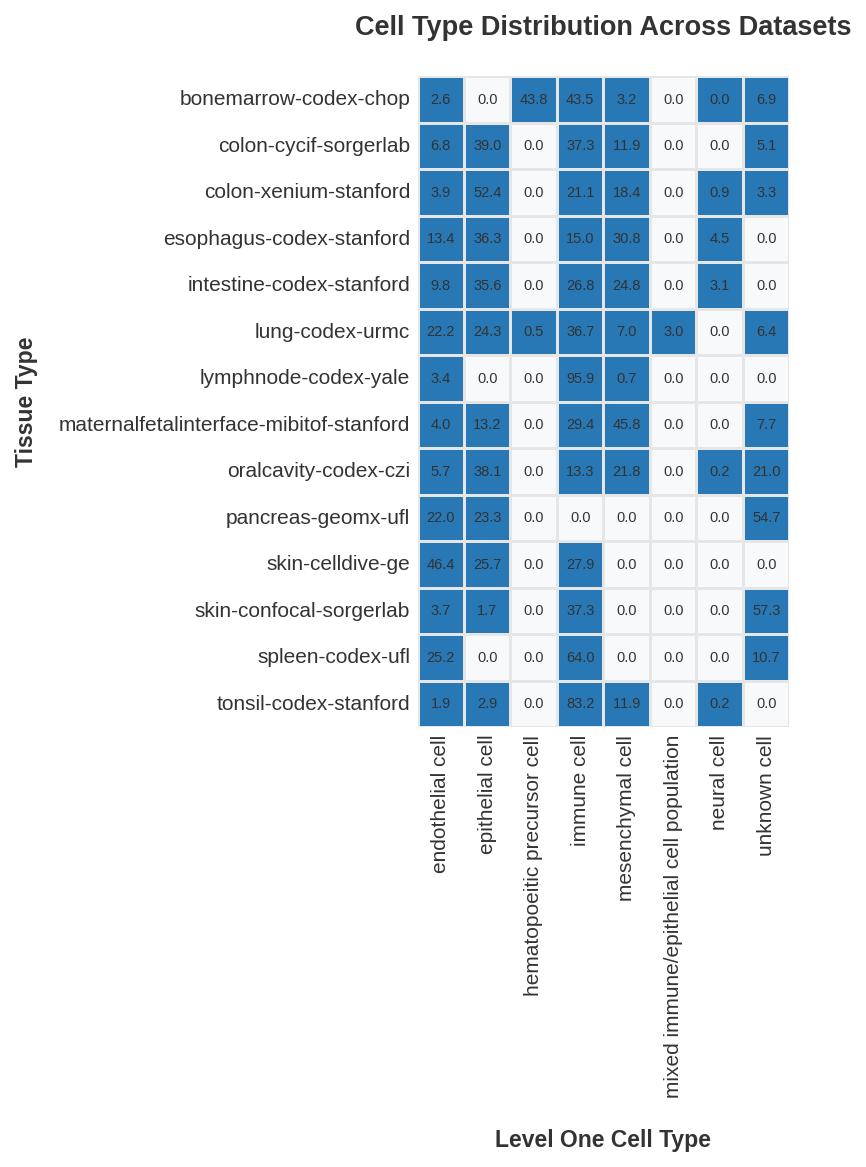
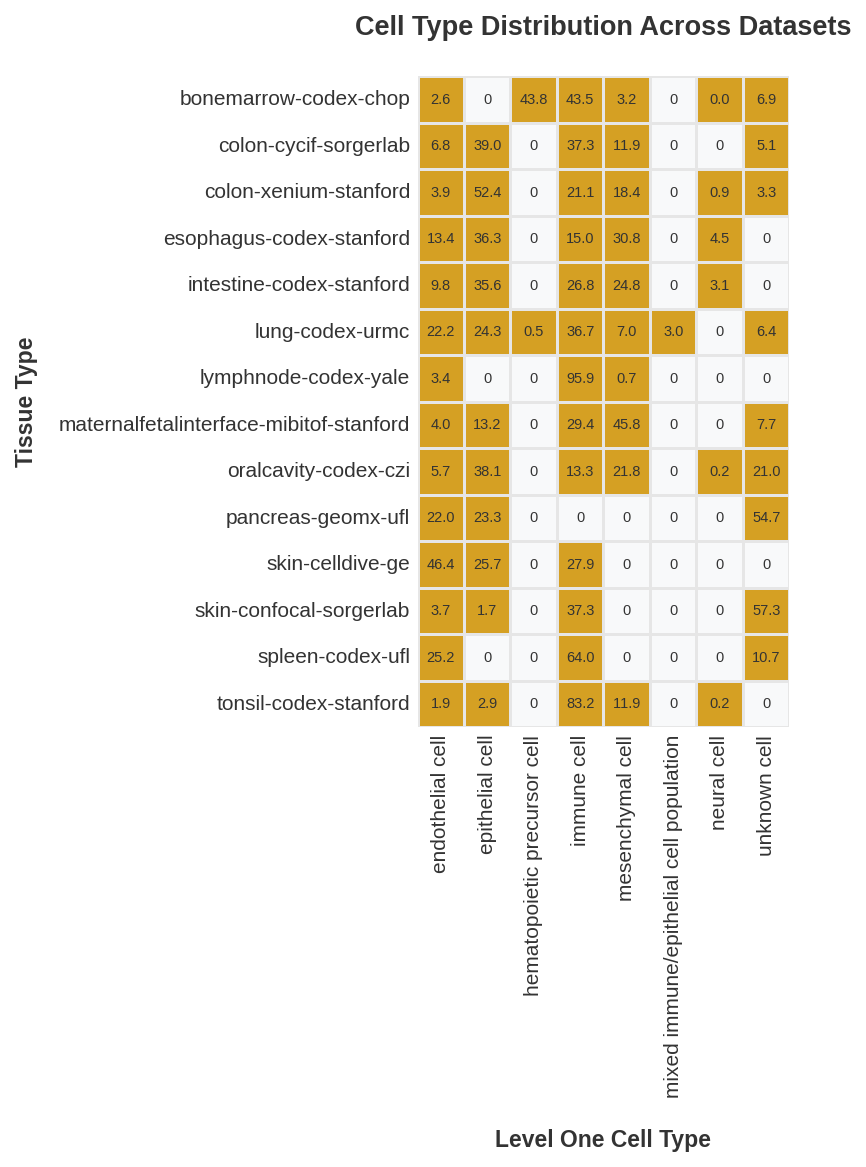
| 0 |
Acinar Cells |
acinar cell of salivary gland |
acinar cell of salivary gland |
CL:0002623 |
skos:exactMatch |
gland epithelium cell |
glandular secretory epithelial cell |
CL:0000150 |
skos:exactMatch |
epithelial cell |
epithelial cell |
CL:0000066 |
skos:exactMatch |
oralcavity-codex-czi |
| 1 |
Acini |
acinar cell of salivary gland |
acinar cell of salivary gland |
CL:0002623 |
skos:exactMatch |
gland epithelium cell |
glandular secretory epithelial cell |
CL:0000150 |
skos:exactMatch |
epithelial cell |
epithelial cell |
CL:0000066 |
skos:exactMatch |
oralcavity-codex-czi |
| 2 |
Adipocytes |
adipocyte |
adipocyte |
CL:0000136 |
skos:exactMatch |
adipocyte |
adipocyte |
CL:0000136 |
skos:exactMatch |
mesenchymal cell |
mesenchymal cell |
CL:0008019 |
skos:exactMatch |
oralcavity-codex-czi |
| 3 |
Adipocytes |
adipocyte |
adipocyte |
CL:0000136 |
skos:exactMatch |
adipocyte |
adipocyte |
CL:0000136 |
skos:exactMatch |
mesenchymal cell |
mesenchymal cell |
CL:0008019 |
skos:exactMatch |
colon-xenium-stanford |
| 4 |
Adipocyte |
adipocyte |
adipocyte |
CL:0000136 |
skos:exactMatch |
adipocyte |
adipocyte |
CL:0000136 |
skos:exactMatch |
mesenchymal cell |
mesenchymal cell |
CL:0008019 |
skos:exactMatch |
bonemarrow-codex-chop |
| 5 |
B-Cells |
b cell |
B cell |
CL:0000236 |
skos:exactMatch |
b cell |
B cell |
CL:0000236 |
skos:exactMatch |
immune cell |
leukocyte |
CL:0000738 |
skos:exactMatch |
bonemarrow-codex-chop |
| 6 |
B |
b cell |
B cell |
CL:0000236 |
skos:exactMatch |
b cell |
B cell |
CL:0000236 |
skos:exactMatch |
immune cell |
leukocyte |
CL:0000738 |
skos:exactMatch |
tonsil-codex-stanford |
| 7 |
B cell |
b cell |
B cell |
CL:0000236 |
skos:exactMatch |
b cell |
B cell |
CL:0000236 |
skos:exactMatch |
immune cell |
leukocyte |
CL:0000738 |
skos:exactMatch |
lung-codex-urmc |
| 8 |
B cells |
b cell |
B cell |
CL:0000236 |
skos:exactMatch |
b cell |
B cell |
CL:0000236 |
skos:exactMatch |
immune cell |
leukocyte |
CL:0000738 |
skos:exactMatch |
colon-cycif-sorgerlab |
| 9 |
B Cells |
b cell |
B cell |
CL:0000236 |
skos:exactMatch |
b cell |
B cell |
CL:0000236 |
skos:exactMatch |
immune cell |
leukocyte |
CL:0000738 |
skos:exactMatch |
oralcavity-codex-czi |
| 10 |
B cells |
b cell |
B cell |
CL:0000236 |
skos:exactMatch |
b cell |
B cell |
CL:0000236 |
skos:exactMatch |
immune cell |
leukocyte |
CL:0000738 |
skos:exactMatch |
skin-confocal-sorgerlab |
| 11 |
B |
b cell |
B cell |
CL:0000236 |
skos:exactMatch |
b cell |
B cell |
CL:0000236 |
skos:exactMatch |
immune cell |
leukocyte |
CL:0000738 |
skos:exactMatch |
intestine-codex-stanford |
| 12 |
B cells, red pulp |
b cell |
B cell |
CL:0000236 |
skos:exactMatch |
b cell |
B cell |
CL:0000236 |
skos:exactMatch |
immune cell |
leukocyte |
CL:0000738 |
skos:exactMatch |
spleen-codex-ufl |
| 13 |
B_cell_1 |
b cell |
B cell |
CL:0000236 |
skos:exactMatch |
b cell |
B cell |
CL:0000236 |
skos:exactMatch |
immune cell |
leukocyte |
CL:0000738 |
skos:exactMatch |
lung-codex-urmc |
| 14 |
B_activated |
b cell:activated |
B cell:activated |
CL:0000236 |
skos:narrowMatch |
b cell |
B cell |
CL:0000236 |
skos:exactMatch |
immune cell |
leukocyte |
CL:0000738 |
skos:exactMatch |
lymphnode-codex-yale |
| 15 |
Fol B cells |
b cell:follicular |
follicular B cell |
CL:0000843 |
skos:exactMatch |
b cell |
B cell |
CL:0000236 |
skos:exactMatch |
immune cell |
leukocyte |
CL:0000738 |
skos:exactMatch |
spleen-codex-ufl |
| 16 |
B_GC_DZ |
b cell:germinal center |
germinal center B cell |
CL:0000844 |
skos:exactMatch |
b cell |
B cell |
CL:0000236 |
skos:exactMatch |
immune cell |
leukocyte |
CL:0000738 |
skos:exactMatch |
lymphnode-codex-yale |
| 17 |
B_GC_LZ |
b cell:germinal center |
germinal center B cell |
CL:0000844 |
skos:exactMatch |
b cell |
B cell |
CL:0000236 |
skos:exactMatch |
immune cell |
leukocyte |
CL:0000738 |
skos:exactMatch |
lymphnode-codex-yale |
| 18 |
B_GC_prePB |
b cell:germinal center pre-plasmablast |
germinal center B cell:pre-plasmablast |
CL:0000844 |
skos:narrowMatch |
b cell |
B cell |
CL:0000236 |
skos:exactMatch |
immune cell |
leukocyte |
CL:0000738 |
skos:exactMatch |
lymphnode-codex-yale |
| 19 |
Immature_B_Cell |
b cell:immature |
immature B cell |
CL:0000816 |
skos:exactMatch |
b cell |
B cell |
CL:0000236 |
skos:exactMatch |
immune cell |
leukocyte |
CL:0000738 |
skos:exactMatch |
bonemarrow-codex-chop |
| 20 |
B_IFN |
b cell:interferon |
B cell:interferon-stimulated |
CL:0000236 |
skos:narrowMatch |
b cell |
B cell |
CL:0000236 |
skos:exactMatch |
immune cell |
leukocyte |
CL:0000738 |
skos:exactMatch |
lymphnode-codex-yale |
| 21 |
B_mem |
b cell:memory |
memory B cell |
CL:0000787 |
skos:exactMatch |
b cell |
B cell |
CL:0000236 |
skos:exactMatch |
immune cell |
leukocyte |
CL:0000738 |
skos:exactMatch |
lymphnode-codex-yale |
| 22 |
Memory B |
b cell:memory |
memory B cell |
CL:0000787 |
skos:exactMatch |
b cell |
B cell |
CL:0000236 |
skos:exactMatch |
immune cell |
leukocyte |
CL:0000738 |
skos:exactMatch |
colon-xenium-stanford |
| 23 |
B_naive |
b cell:naive |
naive B cell |
CL:0000788 |
skos:exactMatch |
b cell |
B cell |
CL:0000236 |
skos:exactMatch |
immune cell |
leukocyte |
CL:0000738 |
skos:exactMatch |
lymphnode-codex-yale |
| 24 |
Naive B |
b cell:naive |
naive B cell |
CL:0000788 |
skos:exactMatch |
b cell |
B cell |
CL:0000236 |
skos:exactMatch |
immune cell |
leukocyte |
CL:0000738 |
skos:exactMatch |
colon-xenium-stanford |
| 25 |
B_preGC |
b cell:pre-germinal center |
B cell:pre-germinal center |
CL:0000236 |
skos:narrowMatch |
b cell |
B cell |
CL:0000236 |
skos:exactMatch |
immune cell |
leukocyte |
CL:0000738 |
skos:exactMatch |
lymphnode-codex-yale |
| 26 |
B_Cycling |
b cell:proliferating |
cycling B cell |
CL:4033068 |
skos:exactMatch |
b cell |
B cell |
CL:0000236 |
skos:exactMatch |
immune cell |
leukocyte |
CL:0000738 |
skos:exactMatch |
lymphnode-codex-yale |
| 27 |
Beta cell |
beta cell:pancreatic |
type B pancreatic cell |
CL:0000169 |
skos:exactMatch |
beta cell |
type B pancreatic cell |
CL:0000169 |
skos:exactMatch |
epithelial cell |
epithelial cell |
CL:0000066 |
skos:exactMatch |
pancreas-geomx-ufl |
| 28 |
CD34+ CD61+ |
cell:cd34+ cd61+ |
leukocyte:cd34-positive cd61-positive |
CL:0000738 |
skos:narrowMatch |
megakaryocyte |
megakaryocyte |
CL:0000556 |
skos:exactMatch |
hematopoietic precursor cell |
hematopoietic precursor cell |
CL:0008001 |
skos:exactMatch |
bonemarrow-codex-chop |
| 29 |
Ki67 proliferating |
cell:proliferating |
cell:proliferating |
CL:0000000 |
skos:narrowMatch |
unknown cell |
cell:unknown |
CL:0000000 |
skos:narrowMatch |
unknown cell |
cell:unknown |
CL:0000000 |
skos:narrowMatch |
spleen-codex-ufl |
| 30 |
DC |
dendritic cell |
dendritic cell |
CL:0000451 |
skos:exactMatch |
dendritic cell |
dendritic cell |
CL:0000451 |
skos:exactMatch |
immune cell |
leukocyte |
CL:0000738 |
skos:exactMatch |
colon-xenium-stanford |
| 31 |
Dendritic cells |
dendritic cell |
dendritic cell |
CL:0000451 |
skos:exactMatch |
dendritic cell |
dendritic cell |
CL:0000451 |
skos:exactMatch |
immune cell |
leukocyte |
CL:0000738 |
skos:exactMatch |
skin-confocal-sorgerlab |
| 32 |
DC |
dendritic cell |
dendritic cell |
CL:0000451 |
skos:exactMatch |
dendritic cell |
dendritic cell |
CL:0000451 |
skos:exactMatch |
immune cell |
leukocyte |
CL:0000738 |
skos:exactMatch |
maternalfetalinterface-mibitof-stanford |
| 33 |
DC |
dendritic cell |
dendritic cell |
CL:0000451 |
skos:exactMatch |
dendritic cell |
dendritic cell |
CL:0000451 |
skos:exactMatch |
immune cell |
leukocyte |
CL:0000738 |
skos:exactMatch |
intestine-codex-stanford |
| 34 |
Dendritic Cells |
dendritic cell |
dendritic cell |
CL:0000451 |
skos:exactMatch |
dendritic cell |
dendritic cell |
CL:0000451 |
skos:exactMatch |
immune cell |
leukocyte |
CL:0000738 |
skos:exactMatch |
oralcavity-codex-czi |
| 35 |
CD11B+ CD11C- cells |
dendritic cell:cd11b+ |
CD11b-positive dendritic cell |
CL:0002465 |
skos:exactMatch |
dendritic cell |
dendritic cell |
CL:0000451 |
skos:exactMatch |
immune cell |
leukocyte |
CL:0000738 |
skos:exactMatch |
skin-confocal-sorgerlab |
| 36 |
DC_cDC1 |
dendritic cell:conventional 1 |
CD141-positive myeloid dendritic cell |
CL:0002394 |
skos:exactMatch |
dendritic cell |
dendritic cell |
CL:0000451 |
skos:exactMatch |
immune cell |
leukocyte |
CL:0000738 |
skos:exactMatch |
lymphnode-codex-yale |
| 37 |
DC_cDC2 |
dendritic cell:conventional 2 |
CD1c-positive myeloid dendritic cell |
CL:0002399 |
skos:exactMatch |
dendritic cell |
dendritic cell |
CL:0000451 |
skos:exactMatch |
immune cell |
leukocyte |
CL:0000738 |
skos:exactMatch |
lymphnode-codex-yale |
| 38 |
FDC |
dendritic cell:follicular |
follicular dendritic cell |
CL:0000442 |
skos:exactMatch |
dendritic cell |
dendritic cell |
CL:0000451 |
skos:exactMatch |
mesenchymal cell |
mesenchymal cell |
CL:0008019 |
skos:exactMatch |
lymphnode-codex-yale |
| 39 |
DC_CCR7+ |
dendritic cell:migratory |
migratory dendtritic cell |
CL:4047054 |
skos:exactMatch |
dendritic cell |
dendritic cell |
CL:0000451 |
skos:exactMatch |
immune cell |
leukocyte |
CL:0000738 |
skos:exactMatch |
lymphnode-codex-yale |
| 40 |
DC_pDC |
dendritic cell:plasmacytoid |
plasmacytoid dendritic cell |
CL:0000784 |
skos:exactMatch |
dendritic cell |
dendritic cell |
CL:0000451 |
skos:exactMatch |
immune cell |
leukocyte |
CL:0000738 |
skos:exactMatch |
lymphnode-codex-yale |
| 41 |
pDC |
dendritic cell:plasmacytoid |
plasmacytoid dendritic cell |
CL:0000784 |
skos:exactMatch |
dendritic cell |
dendritic cell |
CL:0000451 |
skos:exactMatch |
immune cell |
leukocyte |
CL:0000738 |
skos:exactMatch |
bonemarrow-codex-chop |
| 42 |
Endosteal |
endosteal cell |
endosteal cell |
CL:0002157 |
skos:exactMatch |
skeletal stromal cell |
bone cell:stromal |
CL:0001035 |
skos:narrowMatch |
mesenchymal cell |
mesenchymal cell |
CL:0008019 |
skos:exactMatch |
bonemarrow-codex-chop |
| 43 |
Endothelial |
endothelial cell |
endothelial cell |
CL:0000115 |
skos:exactMatch |
endothelial cell |
endothelial cell |
CL:0000115 |
skos:exactMatch |
endothelial cell |
endothelial cell |
CL:0000115 |
skos:exactMatch |
lymphnode-codex-yale |
| 44 |
Endothelial |
endothelial cell |
endothelial cell |
CL:0000115 |
skos:exactMatch |
endothelial cell |
endothelial cell |
CL:0000115 |
skos:exactMatch |
endothelial cell |
endothelial cell |
CL:0000115 |
skos:exactMatch |
skin-celldive-ge |
| 45 |
Endothelial |
endothelial cell |
endothelial cell |
CL:0000115 |
skos:exactMatch |
endothelial cell |
endothelial cell |
CL:0000115 |
skos:exactMatch |
endothelial cell |
endothelial cell |
CL:0000115 |
skos:exactMatch |
skin-confocal-sorgerlab |
| 46 |
Endothelial |
endothelial cell |
endothelial cell |
CL:0000115 |
skos:exactMatch |
endothelial cell |
endothelial cell |
CL:0000115 |
skos:exactMatch |
endothelial cell |
endothelial cell |
CL:0000115 |
skos:exactMatch |
pancreas-geomx-ufl |
| 47 |
Endothelial |
endothelial cell |
endothelial cell |
CL:0000115 |
skos:exactMatch |
endothelial cell |
endothelial cell |
CL:0000115 |
skos:exactMatch |
endothelial cell |
endothelial cell |
CL:0000115 |
skos:exactMatch |
intestine-codex-stanford |
| 48 |
Endo_p |
endothelial cell |
endothelial cell |
CL:0000115 |
skos:exactMatch |
endothelial cell |
endothelial cell |
CL:0000115 |
skos:exactMatch |
endothelial cell |
endothelial cell |
CL:0000115 |
skos:exactMatch |
lung-codex-urmc |
| 49 |
Endothelial |
endothelial cell |
endothelial cell |
CL:0000115 |
skos:exactMatch |
endothelial cell |
endothelial cell |
CL:0000115 |
skos:exactMatch |
endothelial cell |
endothelial cell |
CL:0000115 |
skos:exactMatch |
tonsil-codex-stanford |
| 50 |
Endothelial |
endothelial cell |
endothelial cell |
CL:0000115 |
skos:exactMatch |
endothelial cell |
endothelial cell |
CL:0000115 |
skos:exactMatch |
endothelial cell |
endothelial cell |
CL:0000115 |
skos:exactMatch |
colon-xenium-stanford |
| 51 |
Endothelial |
endothelial cell |
endothelial cell |
CL:0000115 |
skos:exactMatch |
endothelial cell |
endothelial cell |
CL:0000115 |
skos:exactMatch |
endothelial cell |
endothelial cell |
CL:0000115 |
skos:exactMatch |
maternalfetalinterface-mibitof-stanford |
| 52 |
Endothelial |
endothelial cell |
endothelial cell |
CL:0000115 |
skos:exactMatch |
endothelial cell |
endothelial cell |
CL:0000115 |
skos:exactMatch |
endothelial cell |
endothelial cell |
CL:0000115 |
skos:exactMatch |
colon-cycif-sorgerlab |
| 53 |
Endothelial |
endothelial cell |
endothelial cell |
CL:0000115 |
skos:exactMatch |
endothelial cell |
endothelial cell |
CL:0000115 |
skos:exactMatch |
endothelial cell |
endothelial cell |
CL:0000115 |
skos:exactMatch |
esophagus-codex-stanford |
| 54 |
ENDO_SMC |
endothelial cell |
endothelial cell |
CL:0000115 |
skos:exactMatch |
endothelial cell |
endothelial cell |
CL:0000115 |
skos:exactMatch |
endothelial cell |
endothelial cell |
CL:0000115 |
skos:exactMatch |
lung-codex-urmc |
| 55 |
Endo_2 |
endothelial cell |
endothelial cell |
CL:0000115 |
skos:exactMatch |
endothelial cell |
endothelial cell |
CL:0000115 |
skos:exactMatch |
endothelial cell |
endothelial cell |
CL:0000115 |
skos:exactMatch |
lung-codex-urmc |
| 56 |
ENDO_1 |
endothelial cell |
endothelial cell |
CL:0000115 |
skos:exactMatch |
endothelial cell |
endothelial cell |
CL:0000115 |
skos:exactMatch |
endothelial cell |
endothelial cell |
CL:0000115 |
skos:exactMatch |
lung-codex-urmc |
| 57 |
AEC |
endothelial cell of artery |
endothelial cell of artery |
CL:1000413 |
skos:exactMatch |
endothelial cell of artery |
endothelial cell of artery |
CL:1000413 |
skos:exactMatch |
endothelial cell |
endothelial cell |
CL:0000115 |
skos:exactMatch |
bonemarrow-codex-chop |
| 58 |
CAP_ENDO |
endothelial cell of capillary |
capillary endothelial cell |
CL:0002144 |
skos:exactMatch |
endothelial cell of capillary |
capillary endothelial cell |
CL:0002144 |
skos:exactMatch |
endothelial cell |
endothelial cell |
CL:0000115 |
skos:exactMatch |
lung-codex-urmc |
| 59 |
Podoplanin |
endothelial cell of lymphatic vessel |
endothelial cell of lymphatic vessel |
CL:0002138 |
skos:exactMatch |
endothelial cell of lymphatic vessel |
endothelial cell of lymphatic vessel |
CL:0002138 |
skos:exactMatch |
endothelial cell |
endothelial cell |
CL:0000115 |
skos:exactMatch |
spleen-codex-ufl |
| 60 |
Lymphatic endothelial cells |
endothelial cell of lymphatic vessel |
endothelial cell of lymphatic vessel |
CL:0002138 |
skos:exactMatch |
endothelial cell of lymphatic vessel |
endothelial cell of lymphatic vessel |
CL:0002138 |
skos:exactMatch |
endothelial cell |
endothelial cell |
CL:0000115 |
skos:exactMatch |
colon-xenium-stanford |
| 61 |
Lymphatic |
endothelial cell of lymphatic vessel |
endothelial cell of lymphatic vessel |
CL:0002138 |
skos:exactMatch |
endothelial cell of lymphatic vessel |
endothelial cell of lymphatic vessel |
CL:0002138 |
skos:exactMatch |
endothelial cell |
endothelial cell |
CL:0000115 |
skos:exactMatch |
intestine-codex-stanford |
| 62 |
Lymphatic_Endothelium |
endothelial cell of lymphatic vessel |
endothelial cell of lymphatic vessel |
CL:0002138 |
skos:exactMatch |
endothelial cell of lymphatic vessel |
endothelial cell of lymphatic vessel |
CL:0002138 |
skos:exactMatch |
endothelial cell |
endothelial cell |
CL:0000115 |
skos:exactMatch |
lung-codex-urmc |
| 63 |
lymphatic Endothelium |
endothelial cell of lymphatic vessel |
endothelial cell of lymphatic vessel |
CL:0002138 |
skos:exactMatch |
endothelial cell of lymphatic vessel |
endothelial cell of lymphatic vessel |
CL:0002138 |
skos:exactMatch |
endothelial cell |
endothelial cell |
CL:0000115 |
skos:exactMatch |
lung-codex-urmc |
| 64 |
Lymphatic Endothelial Cells |
endothelial cell of lymphatic vessel |
endothelial cell of lymphatic vessel |
CL:0002138 |
skos:exactMatch |
endothelial cell of lymphatic vessel |
endothelial cell of lymphatic vessel |
CL:0002138 |
skos:exactMatch |
endothelial cell |
endothelial cell |
CL:0000115 |
skos:exactMatch |
oralcavity-codex-czi |
| 65 |
Lymphatic Vascular Cells |
endothelial cell of lymphatic vessel |
endothelial cell of lymphatic vessel |
CL:0002138 |
skos:exactMatch |
endothelial cell of lymphatic vessel |
endothelial cell of lymphatic vessel |
CL:0002138 |
skos:exactMatch |
endothelial cell |
endothelial cell |
CL:0000115 |
skos:exactMatch |
oralcavity-codex-czi |
| 66 |
SEC |
endothelial cell of sinusoid |
endothelial cell of sinusoid |
CL:0002262 |
skos:exactMatch |
endothelial cell of sinusoid |
endothelial cell of sinusoid |
CL:0002262 |
skos:exactMatch |
endothelial cell |
endothelial cell |
CL:0000115 |
skos:exactMatch |
bonemarrow-codex-chop |
| 67 |
Sinusoidal cells |
endothelial cell of sinusoid |
endothelial cell of sinusoid |
CL:0002262 |
skos:exactMatch |
endothelial cell of sinusoid |
endothelial cell of sinusoid |
CL:0002262 |
skos:exactMatch |
endothelial cell |
endothelial cell |
CL:0000115 |
skos:exactMatch |
spleen-codex-ufl |
| 68 |
Vascular Endothelial Cells |
endothelial cell of vascular tree |
endothelial cell of vascular tree |
CL:0002139 |
skos:exactMatch |
endothelial cell of vascular tree |
endothelial cell of vascular tree |
CL:0002139 |
skos:exactMatch |
endothelial cell |
endothelial cell |
CL:0000115 |
skos:exactMatch |
oralcavity-codex-czi |
| 69 |
blood endothelial |
endothelial cell of vascular tree |
endothelial cell of vascular tree |
CL:0002139 |
skos:exactMatch |
endothelial cell of vascular tree |
endothelial cell of vascular tree |
CL:0002139 |
skos:exactMatch |
endothelial cell |
endothelial cell |
CL:0000115 |
skos:exactMatch |
spleen-codex-ufl |
| 70 |
VEC Progen |
endothelial cell of vascular tree |
endothelial cell of vascular tree |
CL:0002139 |
skos:exactMatch |
endothelial cell of vascular tree |
endothelial cell of vascular tree |
CL:0002139 |
skos:exactMatch |
endothelial cell |
endothelial cell |
CL:0000115 |
skos:exactMatch |
oralcavity-codex-czi |
| 71 |
Enterocytes |
enterocyte |
enterocyte |
CL:0000584 |
skos:exactMatch |
enterocyte |
enterocyte |
CL:0000584 |
skos:exactMatch |
epithelial cell |
epithelial cell |
CL:0000066 |
skos:exactMatch |
colon-xenium-stanford |
| 72 |
Enterocyte |
enterocyte |
enterocyte |
CL:0000584 |
skos:exactMatch |
enterocyte |
enterocyte |
CL:0000584 |
skos:exactMatch |
epithelial cell |
epithelial cell |
CL:0000066 |
skos:exactMatch |
intestine-codex-stanford |
| 73 |
CD57+ Enterocyte |
enterocyte:cd57+ |
CD57-positive enterocyte |
CL:4033092 |
skos:exactMatch |
enterocyte |
enterocyte |
CL:0000584 |
skos:exactMatch |
epithelial cell |
epithelial cell |
CL:0000066 |
skos:exactMatch |
intestine-codex-stanford |
| 74 |
CD66+ Enterocyte |
enterocyte:cd66+ |
enterocyte:cd66-positive |
CL:0000584 |
skos:narrowMatch |
enterocyte |
enterocyte |
CL:0000584 |
skos:exactMatch |
epithelial cell |
epithelial cell |
CL:0000066 |
skos:exactMatch |
intestine-codex-stanford |
| 75 |
Immature Enterocytes |
enterocyte:immature |
enterocyte:immature |
CL:0000584 |
skos:narrowMatch |
enterocyte |
enterocyte |
CL:0000584 |
skos:exactMatch |
epithelial cell |
epithelial cell |
CL:0000066 |
skos:exactMatch |
colon-xenium-stanford |
| 76 |
MUC1+ Enterocyte |
enterocyte:muc1+ |
enterocyte:muc1-positive |
CL:0000584 |
skos:narrowMatch |
enterocyte |
enterocyte |
CL:0000584 |
skos:exactMatch |
epithelial cell |
epithelial cell |
CL:0000066 |
skos:exactMatch |
intestine-codex-stanford |
| 77 |
Enterocyte Progenitors |
enterocyte:progenitor |
enterocyte:progenitor |
CL:0000584 |
skos:narrowMatch |
enterocyte |
enterocyte |
CL:0000584 |
skos:exactMatch |
epithelial cell |
epithelial cell |
CL:0000066 |
skos:exactMatch |
colon-xenium-stanford |
| 78 |
Enteroendocrine |
enteroendocrine cell |
enteroendocrine cell |
CL:0000164 |
skos:exactMatch |
endocrine cell |
endocrine cell |
CL:0000163 |
skos:exactMatch |
epithelial cell |
epithelial cell |
CL:0000066 |
skos:exactMatch |
colon-xenium-stanford |
| 79 |
Best4+ Enterocytes |
enteroycte:best4+ |
BEST4+ enterocyte |
CL:4030026 |
skos:exactMatch |
enterocyte |
enterocyte |
CL:0000584 |
skos:exactMatch |
epithelial cell |
epithelial cell |
CL:0000066 |
skos:exactMatch |
colon-xenium-stanford |
| 80 |
Epithelial |
epithelial cell |
epithelial cell |
CL:0000066 |
skos:exactMatch |
epithelial cell |
epithelial cell |
CL:0000066 |
skos:exactMatch |
epithelial cell |
epithelial cell |
CL:0000066 |
skos:exactMatch |
oralcavity-codex-czi |
| 81 |
Ionocytes |
epithelial cell |
epithelial cell |
CL:0000066 |
skos:exactMatch |
epithelial cell |
epithelial cell |
CL:0000066 |
skos:exactMatch |
epithelial cell |
epithelial cell |
CL:0000066 |
skos:exactMatch |
oralcavity-codex-czi |
| 82 |
Melanocytes |
epithelial cell |
epithelial cell |
CL:0000066 |
skos:exactMatch |
epithelial cell |
epithelial cell |
CL:0000066 |
skos:exactMatch |
epithelial cell |
epithelial cell |
CL:0000066 |
skos:exactMatch |
oralcavity-codex-czi |
| 83 |
Lung_Epithelial_1 |
epithelial cell |
epithelial cell |
CL:0000066 |
skos:exactMatch |
epithelial cell |
epithelial cell |
CL:0000066 |
skos:exactMatch |
epithelial cell |
epithelial cell |
CL:0000066 |
skos:exactMatch |
lung-codex-urmc |
| 84 |
Lung_Epithelial_4 |
epithelial cell |
epithelial cell |
CL:0000066 |
skos:exactMatch |
epithelial cell |
epithelial cell |
CL:0000066 |
skos:exactMatch |
epithelial cell |
epithelial cell |
CL:0000066 |
skos:exactMatch |
lung-codex-urmc |
| 85 |
Merkel Cells |
epithelial cell |
epithelial cell |
CL:0000066 |
skos:exactMatch |
epithelial cell |
epithelial cell |
CL:0000066 |
skos:exactMatch |
epithelial cell |
epithelial cell |
CL:0000066 |
skos:exactMatch |
oralcavity-codex-czi |
| 86 |
Lung_Epithelial_p |
epithelial cell |
epithelial cell |
CL:0000066 |
skos:exactMatch |
epithelial cell |
epithelial cell |
CL:0000066 |
skos:exactMatch |
epithelial cell |
epithelial cell |
CL:0000066 |
skos:exactMatch |
lung-codex-urmc |
| 87 |
Ducts |
epithelial cell:ductal |
duct epithelial cell |
CL:0000068 |
skos:exactMatch |
gland epithelium cell |
epithelial cell:gland |
CL:0000066 |
skos:narrowMatch |
epithelial cell |
epithelial cell |
CL:0000066 |
skos:exactMatch |
oralcavity-codex-czi |
| 88 |
Ductal Epithelial Cells |
epithelial cell:ductal |
duct epithelial cell |
CL:0000068 |
skos:exactMatch |
gland epithelium cell |
epithelial cell:gland |
CL:0000066 |
skos:narrowMatch |
epithelial cell |
epithelial cell |
CL:0000066 |
skos:exactMatch |
oralcavity-codex-czi |
| 89 |
Ductal cell |
epithelial cell:ductal |
duct epithelial cell |
CL:0000068 |
skos:exactMatch |
gland epithelium cell |
epithelial cell:gland |
CL:0000066 |
skos:narrowMatch |
epithelial cell |
epithelial cell |
CL:0000066 |
skos:exactMatch |
pancreas-geomx-ufl |
| 90 |
Ki67+ Tumor/Epithelial |
epithelial cell:ki67+ proliferating tumor |
epithelial cell:ki67-positive proliferating tumor |
CL:0000066 |
skos:narrowMatch |
abnormal cell |
abnormal cell |
CL:0001061 |
skos:exactMatch |
epithelial cell |
epithelial cell |
CL:0000066 |
skos:exactMatch |
colon-cycif-sorgerlab |
| 91 |
PDL1+ Tumor/Epithelial |
epithelial cell:pdl1+ tumor |
epithelial cell:pdl1-positive tumor |
CL:0000066 |
skos:narrowMatch |
abnormal cell |
abnormal cell |
CL:0001061 |
skos:exactMatch |
epithelial cell |
epithelial cell |
CL:0000066 |
skos:exactMatch |
colon-cycif-sorgerlab |
| 92 |
Secretory_epithelial |
epithelial cell:secretory |
secretory epithelial cell |
CL:1100001 |
skos:exactMatch |
secretory cell of esophagus |
glandular cell of esophagus |
CL:0002657 |
skos:exactMatch |
epithelial cell |
epithelial cell |
CL:0000066 |
skos:exactMatch |
esophagus-codex-stanford |
| 93 |
Erythroblast |
erythroblast |
erythroblast |
CL:0000765 |
skos:exactMatch |
erythroid precursor |
erythroid progenitor cell |
CL:0000038 |
skos:exactMatch |
hematopoietic precursor cell |
hematopoietic precursor cell |
CL:0008001 |
skos:exactMatch |
bonemarrow-codex-chop |
| 94 |
MEP/Early Erythroblast |
erythroblast:basophilic |
basophilic erythroblast |
CL:0000549 |
skos:exactMatch |
erythroid precursor |
erythroid progenitor cell |
CL:0000038 |
skos:exactMatch |
hematopoietic precursor cell |
hematopoietic precursor cell |
CL:0008001 |
skos:exactMatch |
bonemarrow-codex-chop |
| 95 |
Erythroid |
erythroid lineage cell |
erythroid lineage cell |
CL:0000764 |
skos:exactMatch |
erythroid precursor |
erythroid progenitor cell |
CL:0000038 |
skos:exactMatch |
hematopoietic precursor cell |
hematopoietic precursor cell |
CL:0008001 |
skos:exactMatch |
bonemarrow-codex-chop |
| 96 |
Fibroblasts |
fibroblast |
fibroblast |
CL:0000057 |
skos:exactMatch |
fibroblast |
fibroblast |
CL:0000057 |
skos:exactMatch |
mesenchymal cell |
mesenchymal cell |
CL:0008019 |
skos:exactMatch |
oralcavity-codex-czi |
| 97 |
Fibroblasts |
fibroblast |
fibroblast |
CL:0000057 |
skos:exactMatch |
fibroblast |
fibroblast |
CL:0000057 |
skos:exactMatch |
mesenchymal cell |
mesenchymal cell |
CL:0008019 |
skos:exactMatch |
maternalfetalinterface-mibitof-stanford |
| 98 |
Cancer Associated Fibroblasts |
fibroblast:cancer associated |
fibroblast:cancer associated |
CL:0000057 |
skos:narrowMatch |
fibroblast |
fibroblast |
CL:0000057 |
skos:exactMatch |
mesenchymal cell |
mesenchymal cell |
CL:0008019 |
skos:exactMatch |
colon-xenium-stanford |
| 99 |
Crypt Fibroblasts 1 |
fibroblast:crypt 1 |
crypt-top fibroblast:1 |
CL:4052009 |
skos:narrowMatch |
fibroblast |
fibroblast |
CL:0000057 |
skos:exactMatch |
mesenchymal cell |
mesenchymal cell |
CL:0008019 |
skos:exactMatch |
colon-xenium-stanford |
| 100 |
Crypt Fibroblasts 2 |
fibroblast:crypt 2 |
crypt-top fibroblast:2 |
CL:4052009 |
skos:narrowMatch |
fibroblast |
fibroblast |
CL:0000057 |
skos:exactMatch |
mesenchymal cell |
mesenchymal cell |
CL:0008019 |
skos:exactMatch |
colon-xenium-stanford |
| 101 |
Crypt Fibroblasts 3 |
fibroblast:crypt 3 |
crypt-top fibroblast:3 |
CL:4052009 |
skos:narrowMatch |
fibroblast |
fibroblast |
CL:0000057 |
skos:exactMatch |
mesenchymal cell |
mesenchymal cell |
CL:0008019 |
skos:exactMatch |
colon-xenium-stanford |
| 102 |
Crypt Fibroblasts 4 |
fibroblast:crypt 4 |
crypt-top fibroblast:4 |
CL:4052009 |
skos:narrowMatch |
fibroblast |
fibroblast |
CL:0000057 |
skos:exactMatch |
mesenchymal cell |
mesenchymal cell |
CL:0008019 |
skos:exactMatch |
colon-xenium-stanford |
| 103 |
Villus Fibroblasts WNT5B+ |
fibroblast:wnt5b+ villus |
fibroblast:wnt5b+ villus |
CL:0000057 |
skos:narrowMatch |
fibroblast |
fibroblast |
CL:0000057 |
skos:exactMatch |
mesenchymal cell |
mesenchymal cell |
CL:0008019 |
skos:exactMatch |
colon-xenium-stanford |
| 104 |
Glandular_epi |
glandular cell of esophagus |
glandular cell of esophagus |
CL:0002657 |
skos:exactMatch |
gland epithelium cell |
glandular secretory epithelial cell |
CL:0000150 |
skos:exactMatch |
epithelial cell |
epithelial cell |
CL:0000066 |
skos:exactMatch |
esophagus-codex-stanford |
| 105 |
Glandular |
glandular cell of placenta |
glandular secretory epithelial cell:placenta |
CL:0000150 |
skos:narrowMatch |
gland epithelium cell |
glandular secretory epithelial cell |
CL:0000150 |
skos:exactMatch |
epithelial cell |
epithelial cell |
CL:0000066 |
skos:exactMatch |
maternalfetalinterface-mibitof-stanford |
| 106 |
Glia |
glial cell |
glial cell |
CL:0000125 |
skos:exactMatch |
neuroglial cell |
glial cell |
CL:0000125 |
skos:exactMatch |
neural cell |
neural cell |
CL:0002319 |
skos:exactMatch |
colon-xenium-stanford |
| 107 |
Glial/Neuron |
glial cell/neuron |
neural cell:glial/neuron |
CL:0002319 |
skos:narrowMatch |
neuroglial cell/neuron |
neural cell:neuroglial/neuron |
CL:0002319 |
skos:narrowMatch |
neural cell |
neural cell |
CL:0002319 |
skos:exactMatch |
oralcavity-codex-czi |
| 108 |
GC |
goblet cell |
goblet cell |
CL:0000160 |
skos:exactMatch |
goblet cell |
goblet cell |
CL:0000160 |
skos:exactMatch |
epithelial cell |
epithelial cell |
CL:0000066 |
skos:exactMatch |
colon-xenium-stanford |
| 109 |
Goblet |
goblet cell |
goblet cell |
CL:0000160 |
skos:exactMatch |
goblet cell |
goblet cell |
CL:0000160 |
skos:exactMatch |
epithelial cell |
epithelial cell |
CL:0000066 |
skos:exactMatch |
intestine-codex-stanford |
| 110 |
Goblet |
goblet cell |
goblet cell |
CL:0000160 |
skos:exactMatch |
goblet cell |
goblet cell |
CL:0000160 |
skos:exactMatch |
epithelial cell |
epithelial cell |
CL:0000066 |
skos:exactMatch |
colon-xenium-stanford |
| 111 |
Immature Goblet |
goblet cell:immature |
goblet cell:immature |
CL:0000160 |
skos:narrowMatch |
goblet cell |
goblet cell |
CL:0000160 |
skos:exactMatch |
epithelial cell |
epithelial cell |
CL:0000066 |
skos:exactMatch |
colon-xenium-stanford |
| 112 |
GMP |
granulocyte monocyte progenitor cell |
granulocyte monocyte progenitor cell |
CL:0000557 |
skos:exactMatch |
myeloid precursor |
common myeloid progenitor |
CL:0000049 |
skos:exactMatch |
hematopoietic precursor cell |
hematopoietic precursor cell |
CL:0008001 |
skos:exactMatch |
bonemarrow-codex-chop |
| 113 |
GMP/Myeloblast |
granulocyte monocyte progenitor cell/myeloblast |
myeloid lineage restricted progenitor cell:gra... |
CL:0000839 |
skos:narrowMatch |
myeloid precursor |
common myeloid progenitor |
CL:0000049 |
skos:exactMatch |
hematopoietic precursor cell |
hematopoietic precursor cell |
CL:0008001 |
skos:exactMatch |
bonemarrow-codex-chop |
| 114 |
SPINK2+ HSPC |
hematopoietic stem and progenitor cell:spink2+ |
hematopoietic multipotent progenitor cell:spin... |
CL:0000837 |
skos:narrowMatch |
hematopoietic stem and progenitor cell |
hematopoietic precursor cell |
CL:0008001 |
skos:exactMatch |
hematopoietic precursor cell |
hematopoietic precursor cell |
CL:0008001 |
skos:exactMatch |
bonemarrow-codex-chop |
| 115 |
HSC |
hematopoietic stem cell |
hematopoietic stem cell |
CL:0000037 |
skos:exactMatch |
stem cell |
stem cell |
CL:0000034 |
skos:exactMatch |
hematopoietic precursor cell |
hematopoietic precursor cell |
CL:0008001 |
skos:exactMatch |
bonemarrow-codex-chop |
| 116 |
HSPC |
hematopoietic stem cell |
hematopoietic stem cell |
CL:0000037 |
skos:exactMatch |
stem cell |
stem cell |
CL:0000034 |
skos:exactMatch |
hematopoietic precursor cell |
hematopoietic precursor cell |
CL:0008001 |
skos:exactMatch |
bonemarrow-codex-chop |
| 117 |
Placental_Mac |
hofbauer cell |
Hofbauer cell |
CL:3000001 |
skos:exactMatch |
macrophage |
macrophage |
CL:0000235 |
skos:exactMatch |
immune cell |
leukocyte |
CL:0000738 |
skos:exactMatch |
maternalfetalinterface-mibitof-stanford |
| 118 |
B_cell_macrophage_p? |
immune cell |
leukocyte |
CL:0000738 |
skos:exactMatch |
immune cell |
leukocyte |
CL:0000738 |
skos:exactMatch |
immune cell |
leukocyte |
CL:0000738 |
skos:exactMatch |
lung-codex-urmc |
| 119 |
ICC |
interstitial cell of cajal |
interstitial cell of Cajal |
CL:0002088 |
skos:exactMatch |
neurecto-epithelial cell |
neurecto-epithelial cell |
CL:0000710 |
skos:exactMatch |
mesenchymal cell |
mesenchymal cell |
CL:0008019 |
skos:exactMatch |
intestine-codex-stanford |
| 120 |
Keratinocyte |
keratinocyte |
keratinocyte |
CL:0000312 |
skos:exactMatch |
keratinocyte |
keratinocyte |
CL:0000312 |
skos:exactMatch |
epithelial cell |
epithelial cell |
CL:0000066 |
skos:exactMatch |
oralcavity-codex-czi |
| 121 |
Basal Keratincytes |
keratinocyte |
keratinocyte |
CL:0000312 |
skos:exactMatch |
keratinocyte |
keratinocyte |
CL:0000312 |
skos:exactMatch |
epithelial cell |
epithelial cell |
CL:0000066 |
skos:exactMatch |
oralcavity-codex-czi |
| 122 |
Suprabasal Keratinocytes |
keratinocyte |
keratinocyte |
CL:0000312 |
skos:exactMatch |
keratinocyte |
keratinocyte |
CL:0000312 |
skos:exactMatch |
epithelial cell |
epithelial cell |
CL:0000066 |
skos:exactMatch |
oralcavity-codex-czi |
| 123 |
keratinocytes |
keratinocyte |
keratinocyte |
CL:0000312 |
skos:exactMatch |
keratinocyte |
keratinocyte |
CL:0000312 |
skos:exactMatch |
epithelial cell |
epithelial cell |
CL:0000066 |
skos:exactMatch |
skin-confocal-sorgerlab |
| 124 |
DDB2 |
keratinocyte:ddb2+ |
keratinocyte:ddb2-positive |
CL:0000312 |
skos:narrowMatch |
keratinocyte |
keratinocyte |
CL:0000312 |
skos:exactMatch |
epithelial cell |
epithelial cell |
CL:0000066 |
skos:exactMatch |
skin-celldive-ge |
| 125 |
KI67 |
keratinocyte:ki67+ proliferating |
keratinocyte:ki67-positive proliferating |
CL:0000312 |
skos:narrowMatch |
keratinocyte |
keratinocyte |
CL:0000312 |
skos:exactMatch |
epithelial cell |
epithelial cell |
CL:0000066 |
skos:exactMatch |
skin-celldive-ge |
| 126 |
P53 |
keratinocyte:p53+ |
keratinocyte:p53-positive |
CL:0000312 |
skos:narrowMatch |
keratinocyte |
keratinocyte |
CL:0000312 |
skos:exactMatch |
epithelial cell |
epithelial cell |
CL:0000066 |
skos:exactMatch |
skin-celldive-ge |
| 127 |
Langerhan cells |
langerhans cell |
Langerhans cell |
CL:0000453 |
skos:exactMatch |
langerhans cell |
Langerhans cell |
CL:0000453 |
skos:exactMatch |
immune cell |
leukocyte |
CL:0000738 |
skos:exactMatch |
skin-confocal-sorgerlab |
| 128 |
Langerhans Cells |
langerhans cell |
Langerhans cell |
CL:0000453 |
skos:exactMatch |
langerhans cell |
Langerhans cell |
CL:0000453 |
skos:exactMatch |
immune cell |
leukocyte |
CL:0000738 |
skos:exactMatch |
oralcavity-codex-czi |
| 129 |
Leukocyte |
leukocyte |
leukocyte |
CL:0000738 |
skos:exactMatch |
leukocyte |
leukocyte |
CL:0000738 |
skos:exactMatch |
immune cell |
leukocyte |
CL:0000738 |
skos:exactMatch |
lung-codex-urmc |
| 130 |
Other Immune |
leukocyte |
leukocyte |
CL:0000738 |
skos:exactMatch |
leukocyte |
leukocyte |
CL:0000738 |
skos:exactMatch |
immune cell |
leukocyte |
CL:0000738 |
skos:exactMatch |
skin-confocal-sorgerlab |
| 131 |
CD7+ Immune |
lymphocyte:cd7+ |
CD7-positive lymphoid progenitor cell |
CL:0001028 |
skos:exactMatch |
lymphoid cell |
lymphocyte |
CL:0000542 |
skos:exactMatch |
immune cell |
leukocyte |
CL:0000738 |
skos:exactMatch |
intestine-codex-stanford |
| 132 |
DN Lymphocyte |
lymphocyte:double-negative |
double negative thymocyte |
CL:0002489 |
skos:exactMatch |
t cell |
T cell |
CL:0000084 |
skos:exactMatch |
immune cell |
leukocyte |
CL:0000738 |
skos:exactMatch |
colon-cycif-sorgerlab |
| 133 |
DP Lymphocyte |
lymphocyte:double-positive alpha-beta |
double-positive, alpha-beta thymocyte |
CL:0000809 |
skos:exactMatch |
t cell |
T cell |
CL:0000084 |
skos:exactMatch |
immune cell |
leukocyte |
CL:0000738 |
skos:exactMatch |
colon-cycif-sorgerlab |
| 134 |
Lymphocyte(III) |
lymphocyte:iii |
lymphocyte:III |
CL:0000542 |
skos:narrowMatch |
t cell |
T cell |
CL:0000084 |
skos:exactMatch |
immune cell |
leukocyte |
CL:0000738 |
skos:exactMatch |
colon-cycif-sorgerlab |
| 135 |
PDL1+ lymphocyte |
lymphocyte:pdl1+ |
lymphocyte:pdl1-positive |
CL:0000542 |
skos:narrowMatch |
t cell |
T cell |
CL:0000084 |
skos:exactMatch |
immune cell |
leukocyte |
CL:0000738 |
skos:exactMatch |
colon-cycif-sorgerlab |
| 136 |
Innate |
lymphoid cell:innate |
innate lymphoid cell |
CL:0001065 |
skos:exactMatch |
lymphoid cell |
lymphocyte |
CL:0000542 |
skos:exactMatch |
immune cell |
leukocyte |
CL:0000738 |
skos:exactMatch |
tonsil-codex-stanford |
| 137 |
ILC |
lymphoid cell:innate |
innate lymphoid cell |
CL:0001065 |
skos:exactMatch |
lymphoid cell |
lymphocyte |
CL:0000542 |
skos:exactMatch |
immune cell |
leukocyte |
CL:0000738 |
skos:exactMatch |
lymphnode-codex-yale |
| 138 |
Innate |
lymphoid cell:innate |
innate lymphoid cell |
CL:0001065 |
skos:exactMatch |
lymphoid cell |
lymphocyte |
CL:0000542 |
skos:exactMatch |
immune cell |
leukocyte |
CL:0000738 |
skos:exactMatch |
esophagus-codex-stanford |
| 139 |
ILCs |
lymphoid cell:innate |
innate lymphoid cell |
CL:0001065 |
skos:exactMatch |
lymphoid cell |
lymphocyte |
CL:0000542 |
skos:exactMatch |
immune cell |
leukocyte |
CL:0000738 |
skos:exactMatch |
colon-xenium-stanford |
| 140 |
CLP |
lymphoid progenitor cell:common |
common lymphoid progenitor |
CL:0000051 |
skos:exactMatch |
progenitor cell |
hematopoietic precursor cell |
CL:0008001 |
skos:exactMatch |
hematopoietic precursor cell |
hematopoietic precursor cell |
CL:0008001 |
skos:exactMatch |
bonemarrow-codex-chop |
| 141 |
Macrophages_M2 |
macrophage |
macrophage |
CL:0000235 |
skos:exactMatch |
macrophage |
macrophage |
CL:0000235 |
skos:exactMatch |
immune cell |
leukocyte |
CL:0000738 |
skos:exactMatch |
lymphnode-codex-yale |
| 142 |
M2 Macrophage |
macrophage |
macrophage |
CL:0000235 |
skos:exactMatch |
macrophage |
macrophage |
CL:0000235 |
skos:exactMatch |
immune cell |
leukocyte |
CL:0000738 |
skos:exactMatch |
intestine-codex-stanford |
| 143 |
Macrophage |
macrophage |
macrophage |
CL:0000235 |
skos:exactMatch |
macrophage |
macrophage |
CL:0000235 |
skos:exactMatch |
immune cell |
leukocyte |
CL:0000738 |
skos:exactMatch |
oralcavity-codex-czi |
| 144 |
macrophage_CD1c+_myeloidDC |
macrophage |
alveolar macrophage |
CL:0000583 |
skos:exactMatch |
macrophage |
macrophage |
CL:0000235 |
skos:exactMatch |
immune cell |
leukocyte |
CL:0000738 |
skos:exactMatch |
lung-codex-urmc |
| 145 |
Macrophages |
macrophage |
macrophage |
CL:0000235 |
skos:exactMatch |
macrophage |
macrophage |
CL:0000235 |
skos:exactMatch |
immune cell |
leukocyte |
CL:0000738 |
skos:exactMatch |
bonemarrow-codex-chop |
| 146 |
Macrophages |
macrophage |
macrophage |
CL:0000235 |
skos:exactMatch |
macrophage |
macrophage |
CL:0000235 |
skos:exactMatch |
immune cell |
leukocyte |
CL:0000738 |
skos:exactMatch |
colon-xenium-stanford |
| 147 |
CD68 |
macrophage |
macrophage |
CL:0000235 |
skos:exactMatch |
macrophage |
macrophage |
CL:0000235 |
skos:exactMatch |
immune cell |
leukocyte |
CL:0000738 |
skos:exactMatch |
skin-celldive-ge |
| 148 |
Macrophage |
macrophage |
macrophage |
CL:0000235 |
skos:exactMatch |
macrophage |
macrophage |
CL:0000235 |
skos:exactMatch |
immune cell |
leukocyte |
CL:0000738 |
skos:exactMatch |
skin-confocal-sorgerlab |
| 149 |
macrophage |
macrophage |
macrophage |
CL:0000235 |
skos:exactMatch |
macrophage |
macrophage |
CL:0000235 |
skos:exactMatch |
immune cell |
leukocyte |
CL:0000738 |
skos:exactMatch |
lung-codex-urmc |
| 150 |
macrophage_3 |
macrophage |
macrophage |
CL:0000235 |
skos:exactMatch |
macrophage |
macrophage |
CL:0000235 |
skos:exactMatch |
immune cell |
leukocyte |
CL:0000738 |
skos:exactMatch |
lung-codex-urmc |
| 151 |
macrophage_2 |
macrophage |
macrophage |
CL:0000235 |
skos:exactMatch |
macrophage |
macrophage |
CL:0000235 |
skos:exactMatch |
immune cell |
leukocyte |
CL:0000738 |
skos:exactMatch |
lung-codex-urmc |
| 152 |
Macrophages_M1 |
macrophage |
macrophage |
CL:0000235 |
skos:exactMatch |
macrophage |
macrophage |
CL:0000235 |
skos:exactMatch |
immune cell |
leukocyte |
CL:0000738 |
skos:exactMatch |
lymphnode-codex-yale |
| 153 |
Macrophages |
macrophage |
macrophage |
CL:0000235 |
skos:exactMatch |
macrophage |
macrophage |
CL:0000235 |
skos:exactMatch |
immune cell |
leukocyte |
CL:0000738 |
skos:exactMatch |
spleen-codex-ufl |
| 154 |
macrophage_p |
macrophage |
macrophage |
CL:0000235 |
skos:exactMatch |
macrophage |
macrophage |
CL:0000235 |
skos:exactMatch |
immune cell |
leukocyte |
CL:0000738 |
skos:exactMatch |
lung-codex-urmc |
| 155 |
Mac1a |
macrophage:1a |
macrophage:1a |
CL:0000235 |
skos:narrowMatch |
macrophage |
macrophage |
CL:0000235 |
skos:exactMatch |
immune cell |
leukocyte |
CL:0000738 |
skos:exactMatch |
maternalfetalinterface-mibitof-stanford |
| 156 |
Mac1b |
macrophage:1b |
macrophage:1b |
CL:0000235 |
skos:narrowMatch |
macrophage |
macrophage |
CL:0000235 |
skos:exactMatch |
immune cell |
leukocyte |
CL:0000738 |
skos:exactMatch |
maternalfetalinterface-mibitof-stanford |
| 157 |
Mac2a |
macrophage:2a |
macrophage:2a |
CL:0000235 |
skos:narrowMatch |
macrophage |
macrophage |
CL:0000235 |
skos:exactMatch |
immune cell |
leukocyte |
CL:0000738 |
skos:exactMatch |
maternalfetalinterface-mibitof-stanford |
| 158 |
Mac2b |
macrophage:2b |
macrophage:2b |
CL:0000235 |
skos:narrowMatch |
macrophage |
macrophage |
CL:0000235 |
skos:exactMatch |
immune cell |
leukocyte |
CL:0000738 |
skos:exactMatch |
maternalfetalinterface-mibitof-stanford |
| 159 |
Mac2c |
macrophage:2c |
macrophage:2c |
CL:0000235 |
skos:narrowMatch |
macrophage |
macrophage |
CL:0000235 |
skos:exactMatch |
immune cell |
leukocyte |
CL:0000738 |
skos:exactMatch |
maternalfetalinterface-mibitof-stanford |
| 160 |
Macrophage(I) |
macrophage:i |
macrophage:I |
CL:0000235 |
skos:narrowMatch |
macrophage |
macrophage |
CL:0000235 |
skos:exactMatch |
immune cell |
leukocyte |
CL:0000738 |
skos:exactMatch |
colon-cycif-sorgerlab |
| 161 |
Macrophage(II) |
macrophage:ii |
macrophage:II |
CL:0000235 |
skos:narrowMatch |
macrophage |
macrophage |
CL:0000235 |
skos:exactMatch |
immune cell |
leukocyte |
CL:0000738 |
skos:exactMatch |
colon-cycif-sorgerlab |
| 162 |
Macrophage(III) |
macrophage:iii |
macrophage:III |
CL:0000235 |
skos:narrowMatch |
macrophage |
macrophage |
CL:0000235 |
skos:exactMatch |
immune cell |
leukocyte |
CL:0000738 |
skos:exactMatch |
colon-cycif-sorgerlab |
| 163 |
M1 Macrophage |
macrophage:inflammatory |
inflammatory macrophage |
CL:0000863 |
skos:exactMatch |
macrophage |
macrophage |
CL:0000235 |
skos:exactMatch |
immune cell |
leukocyte |
CL:0000738 |
skos:exactMatch |
intestine-codex-stanford |
| 164 |
lung Intersitial macrophage |
macrophage:interstitial |
lung interstitial macrophage |
CL:4033043 |
skos:narrowMatch |
macrophage |
macrophage |
CL:0000235 |
skos:exactMatch |
immune cell |
leukocyte |
CL:0000738 |
skos:exactMatch |
lung-codex-urmc |
| 165 |
Macrophage(IV) |
macrophage:iv |
macrophage:IV |
CL:0000235 |
skos:narrowMatch |
macrophage |
macrophage |
CL:0000235 |
skos:exactMatch |
immune cell |
leukocyte |
CL:0000738 |
skos:exactMatch |
colon-cycif-sorgerlab |
| 166 |
PDL1+ Macrophage |
macrophage:pdl1+ |
macrophage:pdl1-positive |
CL:0000235 |
skos:narrowMatch |
macrophage |
macrophage |
CL:0000235 |
skos:exactMatch |
immune cell |
leukocyte |
CL:0000738 |
skos:exactMatch |
colon-cycif-sorgerlab |
| 167 |
Mast |
mast cell |
mast cell |
CL:0000097 |
skos:exactMatch |
mast cell |
mast cell |
CL:0000097 |
skos:exactMatch |
immune cell |
leukocyte |
CL:0000738 |
skos:exactMatch |
lymphnode-codex-yale |
| 168 |
Mast Cells |
mast cell |
mast cell |
CL:0000097 |
skos:exactMatch |
mast cell |
mast cell |
CL:0000097 |
skos:exactMatch |
immune cell |
leukocyte |
CL:0000738 |
skos:exactMatch |
oralcavity-codex-czi |
| 169 |
Mast |
mast cell |
mast cell |
CL:0000097 |
skos:exactMatch |
mast cell |
mast cell |
CL:0000097 |
skos:exactMatch |
immune cell |
leukocyte |
CL:0000738 |
skos:exactMatch |
colon-xenium-stanford |
| 170 |
Mast |
mast cell |
mast cell |
CL:0000097 |
skos:exactMatch |
mast cell |
mast cell |
CL:0000097 |
skos:exactMatch |
immune cell |
leukocyte |
CL:0000738 |
skos:exactMatch |
maternalfetalinterface-mibitof-stanford |
| 171 |
Mast cell |
mast cell |
mast cell |
CL:0000097 |
skos:exactMatch |
mast cell |
mast cell |
CL:0000097 |
skos:exactMatch |
immune cell |
leukocyte |
CL:0000738 |
skos:exactMatch |
lung-codex-urmc |
| 172 |
megakaryocyte |
megakaryocyte |
megakaryocyte |
CL:0000556 |
skos:exactMatch |
megakaryocyte |
megakaryocyte |
CL:0000556 |
skos:exactMatch |
hematopoietic precursor cell |
hematopoietic precursor cell |
CL:0008001 |
skos:exactMatch |
lung-codex-urmc |
| 173 |
GATA1pos_Mks |
megakaryocyte:gata1+ |
megakaryocyte:gata1-positive |
CL:0000556 |
skos:narrowMatch |
megakaryocyte |
megakaryocyte |
CL:0000556 |
skos:exactMatch |
hematopoietic precursor cell |
hematopoietic precursor cell |
CL:0008001 |
skos:exactMatch |
bonemarrow-codex-chop |
| 174 |
GATA1neg_Mks |
megakaryocyte:gata1- |
megakaryocyte:gata1-negative |
CL:0000556 |
skos:narrowMatch |
megakaryocyte |
megakaryocyte |
CL:0000556 |
skos:exactMatch |
hematopoietic precursor cell |
hematopoietic precursor cell |
CL:0008001 |
skos:exactMatch |
bonemarrow-codex-chop |
| 175 |
Adipo-MSC |
mesenchymal stem cell of adipose tissue |
mesenchymal stem cell of adipose tissue |
CL:0002570 |
skos:exactMatch |
mesenchymal stem cell |
mesenchymal stem cell |
CL:0000134 |
skos:exactMatch |
mesenchymal cell |
mesenchymal cell |
CL:0008019 |
skos:exactMatch |
bonemarrow-codex-chop |
| 176 |
THY1+ MSC |
mesenchymal stem/stromal cell:thy1+ |
mesenchymal stem cell:thy1-positive |
CL:0000134 |
skos:narrowMatch |
mesenchymal stem/stromal cell |
mesenchymal stem cell |
CL:0000134 |
skos:exactMatch |
hematopoietic precursor cell |
hematopoietic precursor cell |
CL:0008001 |
skos:exactMatch |
bonemarrow-codex-chop |
| 177 |
Lung_Epithelil_2_CD4+_T_cell |
mixed t cell/epithelial cell population |
cell:mixed t cell/epithelial cell population |
CL:0000000 |
skos:narrowMatch |
mixed t cell/epithelial cell population |
T cell/epithelial cell |
CL:0000084/CL:0000066 |
skos:narrowMatch |
mixed immune/epithelial cell population |
T cell/epithelial cell |
CL:0000084/CL:0000066 |
skos:narrowMatch |
lung-codex-urmc |
| 178 |
Monocytes |
monocyte |
monocyte |
CL:0000576 |
skos:exactMatch |
monocyte |
monocyte |
CL:0000576 |
skos:exactMatch |
immune cell |
leukocyte |
CL:0000738 |
skos:exactMatch |
lymphnode-codex-yale |
| 179 |
Monocytes |
monocyte |
monocyte |
CL:0000576 |
skos:exactMatch |
monocyte |
monocyte |
CL:0000576 |
skos:exactMatch |
immune cell |
leukocyte |
CL:0000738 |
skos:exactMatch |
bonemarrow-codex-chop |
| 180 |
Monocyte-Macrophage |
monocyte |
monocyte |
CL:0000576 |
skos:exactMatch |
macrophage |
macrophage |
CL:0000235 |
skos:exactMatch |
immune cell |
leukocyte |
CL:0000738 |
skos:exactMatch |
oralcavity-codex-czi |
| 181 |
Non-Classical Monocyte |
monocyte:non-classical |
non-classical monocyte |
CL:0000875 |
skos:exactMatch |
monocyte |
monocyte |
CL:0000576 |
skos:exactMatch |
immune cell |
leukocyte |
CL:0000738 |
skos:exactMatch |
bonemarrow-codex-chop |
| 182 |
Mural Cells |
mural cell |
mural cell |
CL:0008034 |
skos:exactMatch |
perivascular cell |
perivascular cell |
CL:4033054 |
skos:exactMatch |
mesenchymal cell |
mesenchymal cell |
CL:0008019 |
skos:exactMatch |
oralcavity-codex-czi |
| 183 |
muscle |
muscle cell |
muscle cell |
CL:0000187 |
skos:exactMatch |
muscle cell |
muscle cell |
CL:0000187 |
skos:exactMatch |
mesenchymal cell |
mesenchymal cell |
CL:0008019 |
skos:exactMatch |
maternalfetalinterface-mibitof-stanford |
| 184 |
Skeletal Myocytes |
muscle cell:skeletal |
cell of skeletal muscle |
CL:0000188 |
skos:exactMatch |
muscle cell |
muscle cell |
CL:0000187 |
skos:exactMatch |
mesenchymal cell |
mesenchymal cell |
CL:0008019 |
skos:exactMatch |
oralcavity-codex-czi |
| 185 |
Smooth muscle |
muscle cell:smooth |
smooth muscle cell |
CL:0000192 |
skos:exactMatch |
muscle cell |
muscle cell |
CL:0000187 |
skos:exactMatch |
mesenchymal cell |
mesenchymal cell |
CL:0008019 |
skos:exactMatch |
intestine-codex-stanford |
| 186 |
SMC_1 |
muscle cell:smooth |
smooth muscle cell |
CL:0000192 |
skos:exactMatch |
muscle cell |
muscle cell |
CL:0000187 |
skos:exactMatch |
mesenchymal cell |
mesenchymal cell |
CL:0008019 |
skos:exactMatch |
lung-codex-urmc |
| 187 |
VSMC |
muscle cell:smooth |
smooth muscle cell |
CL:0000192 |
skos:exactMatch |
muscle cell |
muscle cell |
CL:0000187 |
skos:exactMatch |
mesenchymal cell |
mesenchymal cell |
CL:0008019 |
skos:exactMatch |
lymphnode-codex-yale |
| 188 |
SMC_2 |
muscle cell:smooth |
smooth muscle cell |
CL:0000192 |
skos:exactMatch |
muscle cell |
muscle cell |
CL:0000187 |
skos:exactMatch |
mesenchymal cell |
mesenchymal cell |
CL:0008019 |
skos:exactMatch |
lung-codex-urmc |
| 189 |
SmoothMuscle |
muscle cell:smooth |
smooth muscle cell |
CL:0000192 |
skos:exactMatch |
muscle cell |
muscle cell |
CL:0000187 |
skos:exactMatch |
mesenchymal cell |
mesenchymal cell |
CL:0008019 |
skos:exactMatch |
tonsil-codex-stanford |
| 190 |
smooth muscle_2 |
muscle cell:smooth |
smooth muscle cell |
CL:0000192 |
skos:exactMatch |
muscle cell |
muscle cell |
CL:0000187 |
skos:exactMatch |
mesenchymal cell |
mesenchymal cell |
CL:0008019 |
skos:exactMatch |
lung-codex-urmc |
| 191 |
SmoothMuscle |
muscle cell:smooth |
smooth muscle cell of the esophagus |
CL:0002599 |
skos:exactMatch |
muscle cell |
muscle cell |
CL:0000187 |
skos:exactMatch |
mesenchymal cell |
mesenchymal cell |
CL:0008019 |
skos:exactMatch |
esophagus-codex-stanford |
| 192 |
VSMC |
muscle cell:smooth |
smooth muscle cell |
CL:0000192 |
skos:exactMatch |
muscle cell |
muscle cell |
CL:0000187 |
skos:exactMatch |
mesenchymal cell |
mesenchymal cell |
CL:0008019 |
skos:exactMatch |
bonemarrow-codex-chop |
| 193 |
smooth muscle 1 |
muscle cell:smooth |
smooth muscle cell |
CL:0000192 |
skos:exactMatch |
muscle cell |
muscle cell |
CL:0000187 |
skos:exactMatch |
mesenchymal cell |
mesenchymal cell |
CL:0008019 |
skos:exactMatch |
lung-codex-urmc |
| 194 |
NPM1 Mutant Blast |
mutant blast:npm1 |
NaN |
NaN |
NaN |
abnormal cell |
abnormal cell |
CL:0001061 |
skos:exactMatch |
hematopoietic precursor cell |
hematopoietic precursor cell |
CL:0008001 |
skos:exactMatch |
bonemarrow-codex-chop |
| 195 |
Myeloid |
myeloid cell |
myeloid cell |
CL:0000763 |
skos:exactMatch |
myeloid cell |
myeloid cell |
CL:0000763 |
skos:exactMatch |
immune cell |
leukocyte |
CL:0000738 |
skos:exactMatch |
skin-confocal-sorgerlab |
| 196 |
Myeloid cells |
myeloid cell |
myeloid cell |
CL:0000763 |
skos:exactMatch |
myeloid cell |
myeloid cell |
CL:0000763 |
skos:exactMatch |
immune cell |
leukocyte |
CL:0000738 |
skos:exactMatch |
spleen-codex-ufl |
| 197 |
Intermediate Myeloid |
myeloid cell:intermediate |
myeloid cell:intermediate |
CL:0000763 |
skos:narrowMatch |
myeloid precursor |
common myeloid progenitor |
CL:0000049 |
skos:exactMatch |
hematopoietic precursor cell |
hematopoietic precursor cell |
CL:0008001 |
skos:exactMatch |
bonemarrow-codex-chop |
| 198 |
Mature Myeloid |
myeloid cell:mature |
myeloid cell:mature |
CL:0000763 |
skos:narrowMatch |
myeloid cell |
myeloid cell |
CL:0000763 |
skos:exactMatch |
immune cell |
leukocyte |
CL:0000738 |
skos:exactMatch |
bonemarrow-codex-chop |
| 199 |
Early Myeloid Progenitor |
myeloid progenitor cell:common |
common myeloid progenitor |
CL:0000049 |
skos:exactMatch |
progenitor cell |
hematopoietic precursor cell |
CL:0008001 |
skos:exactMatch |
hematopoietic precursor cell |
hematopoietic precursor cell |
CL:0008001 |
skos:exactMatch |
bonemarrow-codex-chop |
| 200 |
Myoepithelial Cells |
myoepithelial cell |
myoepithelial cell |
CL:0000185 |
skos:exactMatch |
myoepithelial cell |
myoepithelial cell |
CL:0000185 |
skos:exactMatch |
epithelial cell |
epithelial cell |
CL:0000066 |
skos:exactMatch |
oralcavity-codex-czi |
| 201 |
Muscle/Fibroblast |
myofibroblast |
myofibroblast cell |
CL:0000186 |
skos:exactMatch |
fibroblast |
fibroblast |
CL:0000057 |
skos:exactMatch |
mesenchymal cell |
mesenchymal cell |
CL:0008019 |
skos:exactMatch |
colon-cycif-sorgerlab |
| 202 |
Myofibroblasts |
myofibroblast |
myofibroblast cell |
CL:0000186 |
skos:exactMatch |
fibroblast |
fibroblast |
CL:0000057 |
skos:exactMatch |
mesenchymal cell |
mesenchymal cell |
CL:0008019 |
skos:exactMatch |
maternalfetalinterface-mibitof-stanford |
| 203 |
myofibroblast |
myofibroblast |
myofibroblast cell |
CL:0000186 |
skos:exactMatch |
fibroblast |
fibroblast |
CL:0000057 |
skos:exactMatch |
mesenchymal cell |
mesenchymal cell |
CL:0008019 |
skos:exactMatch |
oralcavity-codex-czi |
| 204 |
Myofibroblasts/Smooth Muscle 1 |
myofibroblast cell:smooth muscle 1 |
myofibroblast cell:smooth muscle-like 1 |
CL:0000186 |
skos:narrowMatch |
fibroblast |
fibroblast |
CL:0000057 |
skos:exactMatch |
mesenchymal cell |
mesenchymal cell |
CL:0008019 |
skos:exactMatch |
colon-xenium-stanford |
| 205 |
Myofibroblasts/Smooth Muscle 2 |
myofibroblast cell:smooth muscle 2 |
myofibroblast cell:smooth muscle-like 2 |
CL:0000186 |
skos:narrowMatch |
fibroblast |
fibroblast |
CL:0000057 |
skos:exactMatch |
mesenchymal cell |
mesenchymal cell |
CL:0008019 |
skos:exactMatch |
colon-xenium-stanford |
| 206 |
Myofibroblasts/Smooth Muscle 3 |
myofibroblast cell:smooth muscle 3 |
myofibroblast cell:smooth muscle-like 3 |
CL:0000186 |
skos:narrowMatch |
fibroblast |
fibroblast |
CL:0000057 |
skos:exactMatch |
mesenchymal cell |
mesenchymal cell |
CL:0008019 |
skos:exactMatch |
colon-xenium-stanford |
| 207 |
NK |
natural killer cell |
natural killer cell |
CL:0000623 |
skos:exactMatch |
natural killer cell |
natural killer cell |
CL:0000623 |
skos:exactMatch |
immune cell |
leukocyte |
CL:0000738 |
skos:exactMatch |
lymphnode-codex-yale |
| 208 |
NK1 |
natural killer cell |
natural killer cell |
CL:0000623 |
skos:exactMatch |
natural killer cell |
natural killer cell |
CL:0000623 |
skos:exactMatch |
immune cell |
leukocyte |
CL:0000738 |
skos:exactMatch |
maternalfetalinterface-mibitof-stanford |
| 209 |
NK2 |
natural killer cell |
natural killer cell |
CL:0000623 |
skos:exactMatch |
natural killer cell |
natural killer cell |
CL:0000623 |
skos:exactMatch |
immune cell |
leukocyte |
CL:0000738 |
skos:exactMatch |
maternalfetalinterface-mibitof-stanford |
| 210 |
NK3 |
natural killer cell |
natural killer cell |
CL:0000623 |
skos:exactMatch |
natural killer cell |
natural killer cell |
CL:0000623 |
skos:exactMatch |
immune cell |
leukocyte |
CL:0000738 |
skos:exactMatch |
maternalfetalinterface-mibitof-stanford |
| 211 |
NK4 |
natural killer cell |
natural killer cell |
CL:0000623 |
skos:exactMatch |
natural killer cell |
natural killer cell |
CL:0000623 |
skos:exactMatch |
immune cell |
leukocyte |
CL:0000738 |
skos:exactMatch |
maternalfetalinterface-mibitof-stanford |
| 212 |
NK Cells |
natural killer cell |
natural killer cell |
CL:0000623 |
skos:exactMatch |
natural killer cell |
natural killer cell |
CL:0000623 |
skos:exactMatch |
immune cell |
leukocyte |
CL:0000738 |
skos:exactMatch |
oralcavity-codex-czi |
| 213 |
NK |
natural killer cell |
natural killer cell |
CL:0000623 |
skos:exactMatch |
natural killer cell |
natural killer cell |
CL:0000623 |
skos:exactMatch |
immune cell |
leukocyte |
CL:0000738 |
skos:exactMatch |
intestine-codex-stanford |
| 214 |
Tumor/Epithelial |
neoplastic cell |
neoplastic cell |
CL:0001063 |
skos:exactMatch |
abnormal cell |
abnormal cell |
CL:0001061 |
skos:exactMatch |
epithelial cell |
epithelial cell |
CL:0000066 |
skos:exactMatch |
colon-cycif-sorgerlab |
| 215 |
Neuroendocrine |
neuroendocrine cell |
neuroendocrine cell |
CL:0000165 |
skos:exactMatch |
endocrine cell |
endocrine cell |
CL:0000163 |
skos:exactMatch |
epithelial cell |
epithelial cell |
CL:0000066 |
skos:exactMatch |
intestine-codex-stanford |
| 216 |
Nerve |
neuron |
neuron |
CL:0000540 |
skos:exactMatch |
neuron |
neuron |
CL:0000540 |
skos:exactMatch |
neural cell |
neural cell |
CL:0002319 |
skos:exactMatch |
esophagus-codex-stanford |
| 217 |
Neurons |
neuron |
neuron |
CL:0000540 |
skos:exactMatch |
neuron |
neuron |
CL:0000540 |
skos:exactMatch |
neural cell |
neural cell |
CL:0002319 |
skos:exactMatch |
colon-xenium-stanford |
| 218 |
Nerve |
neuron |
neuron |
CL:0000540 |
skos:exactMatch |
neuron |
neuron |
CL:0000540 |
skos:exactMatch |
neural cell |
neural cell |
CL:0002319 |
skos:exactMatch |
tonsil-codex-stanford |
| 219 |
Nerve |
neuron |
neuron |
CL:0000540 |
skos:exactMatch |
neuron |
neuron |
CL:0000540 |
skos:exactMatch |
neural cell |
neural cell |
CL:0002319 |
skos:exactMatch |
intestine-codex-stanford |
| 220 |
Neutrophil |
neutrophil |
neutrophil |
CL:0000775 |
skos:exactMatch |
neutrophil |
neutrophil |
CL:0000775 |
skos:exactMatch |
immune cell |
leukocyte |
CL:0000738 |
skos:exactMatch |
intestine-codex-stanford |
| 221 |
neutrophil_2 |
neutrophil |
neutrophil |
CL:0000775 |
skos:exactMatch |
neutrophil |
neutrophil |
CL:0000775 |
skos:exactMatch |
immune cell |
leukocyte |
CL:0000738 |
skos:exactMatch |
lung-codex-urmc |
| 222 |
Neutrophils |
neutrophil |
neutrophil |
CL:0000775 |
skos:exactMatch |
neutrophil |
neutrophil |
CL:0000775 |
skos:exactMatch |
immune cell |
leukocyte |
CL:0000738 |
skos:exactMatch |
oralcavity-codex-czi |
| 223 |
neutrophil |
neutrophil |
neutrophil |
CL:0000775 |
skos:exactMatch |
neutrophil |
neutrophil |
CL:0000775 |
skos:exactMatch |
immune cell |
leukocyte |
CL:0000738 |
skos:exactMatch |
lung-codex-urmc |
| 224 |
Neutrophils/Monocytes |
neutrophil/monocyte |
myeloid leukocyte:neutrophil/monocyte |
CL:0000766 |
skos:narrowMatch |
neutrophil |
neutrophil |
CL:0000775 |
skos:exactMatch |
immune cell |
leukocyte |
CL:0000738 |
skos:exactMatch |
spleen-codex-ufl |
| 225 |
Paneth |
paneth cell |
paneth cell |
CL:0000510 |
skos:exactMatch |
paneth cell |
paneth cell |
CL:0000510 |
skos:exactMatch |
epithelial cell |
epithelial cell |
CL:0000066 |
skos:exactMatch |
intestine-codex-stanford |
| 226 |
Paneth |
paneth cell |
paneth cell |
CL:0000510 |
skos:exactMatch |
paneth cell |
paneth cell |
CL:0000510 |
skos:exactMatch |
epithelial cell |
epithelial cell |
CL:0000066 |
skos:exactMatch |
esophagus-codex-stanford |
| 227 |
Pericytes |
pericyte |
pericyte |
CL:0000669 |
skos:exactMatch |
pericyte |
pericyte |
CL:0000669 |
skos:exactMatch |
mesenchymal cell |
mesenchymal cell |
CL:0008019 |
skos:exactMatch |
colon-xenium-stanford |
| 228 |
Plasma Cells |
plasma cell |
plasma cell |
CL:0000786 |
skos:exactMatch |
b cell |
B cell |
CL:0000236 |
skos:exactMatch |
immune cell |
leukocyte |
CL:0000738 |
skos:exactMatch |
bonemarrow-codex-chop |
| 229 |
B_plasma |
plasma cell |
plasma cell |
CL:0000786 |
skos:exactMatch |
b cell |
B cell |
CL:0000236 |
skos:exactMatch |
immune cell |
leukocyte |
CL:0000738 |
skos:exactMatch |
lymphnode-codex-yale |
| 230 |
Plasma |
plasma cell |
plasma cell |
CL:0000786 |
skos:exactMatch |
b cell |
B cell |
CL:0000236 |
skos:exactMatch |
immune cell |
leukocyte |
CL:0000738 |
skos:exactMatch |
tonsil-codex-stanford |
| 231 |
Plasma |
plasma cell |
plasma cell |
CL:0000786 |
skos:exactMatch |
b cell |
B cell |
CL:0000236 |
skos:exactMatch |
immune cell |
leukocyte |
CL:0000738 |
skos:exactMatch |
colon-xenium-stanford |
| 232 |
Plasma Cells |
plasma cell |
plasma cell |
CL:0000786 |
skos:exactMatch |
b cell |
B cell |
CL:0000236 |
skos:exactMatch |
immune cell |
leukocyte |
CL:0000738 |
skos:exactMatch |
oralcavity-codex-czi |
| 233 |
Plasma |
plasma cell |
plasma cell |
CL:0000786 |
skos:exactMatch |
b cell |
B cell |
CL:0000236 |
skos:exactMatch |
immune cell |
leukocyte |
CL:0000738 |
skos:exactMatch |
intestine-codex-stanford |
| 234 |
Plasma |
plasma cell |
plasma cell |
CL:0000786 |
skos:exactMatch |
b cell |
B cell |
CL:0000236 |
skos:exactMatch |
immune cell |
leukocyte |
CL:0000738 |
skos:exactMatch |
esophagus-codex-stanford |
| 235 |
AT1_1 |
pneumocyte:type 1 |
pulmonary alveolar type 1 cell |
CL:0002062 |
skos:exactMatch |
type 1 pneumocyte |
pulmonary alveolar type 1 cell |
CL:0002062 |
skos:exactMatch |
epithelial cell |
epithelial cell |
CL:0000066 |
skos:exactMatch |
lung-codex-urmc |
| 236 |
AT1 _2 |
pneumocyte:type 1 |
pulmonary alveolar type 1 cell |
CL:0002062 |
skos:exactMatch |
type 1 pneumocyte |
pulmonary alveolar type 1 cell |
CL:0002062 |
skos:exactMatch |
epithelial cell |
epithelial cell |
CL:0000066 |
skos:exactMatch |
lung-codex-urmc |
| 237 |
AT2_p |
pneumocyte:type 2 |
pulmonary alveolar type 2 cell |
CL:0002063 |
skos:exactMatch |
type 2 pneumocyte |
pulmonary alveolar type 2 cell |
CL:0002063 |
skos:exactMatch |
epithelial cell |
epithelial cell |
CL:0000066 |
skos:exactMatch |
lung-codex-urmc |
| 238 |
AT2_2 |
pneumocyte:type 2 |
pulmonary alveolar type 2 cell |
CL:0002063 |
skos:exactMatch |
type 2 pneumocyte |
pulmonary alveolar type 2 cell |
CL:0002063 |
skos:exactMatch |
epithelial cell |
epithelial cell |
CL:0000066 |
skos:exactMatch |
lung-codex-urmc |
| 239 |
AT2 |
pneumocyte:type 2 |
pulmonary alveolar type 2 cell |
CL:0002063 |
skos:exactMatch |
type 2 pneumocyte |
pulmonary alveolar type 2 cell |
CL:0002063 |
skos:exactMatch |
epithelial cell |
epithelial cell |
CL:0000066 |
skos:exactMatch |
lung-codex-urmc |
| 240 |
AT2_1 |
pneumocyte:type 2 |
pulmonary alveolar type 2 cell |
CL:0002063 |
skos:exactMatch |
type 2 pneumocyte |
pulmonary alveolar type 2 cell |
CL:0002063 |
skos:exactMatch |
epithelial cell |
epithelial cell |
CL:0000066 |
skos:exactMatch |
lung-codex-urmc |
| 241 |
Schwann Cells |
schwann cell |
Schwann cell |
CL:0002573 |
skos:exactMatch |
neuroglial cell |
glial cell |
CL:0000125 |
skos:exactMatch |
neural cell |
neural cell |
CL:0002319 |
skos:exactMatch |
bonemarrow-codex-chop |
| 242 |
Squamous_epithelial |
squamous epithelial cell |
squamous epithelial cell |
CL:0000076 |
skos:exactMatch |
squamous epithelial cell |
squamous epithelial cell |
CL:0000076 |
skos:exactMatch |
epithelial cell |
epithelial cell |
CL:0000066 |
skos:exactMatch |
esophagus-codex-stanford |
| 243 |
Squamous_epithelial |
squamous epithelial cell |
squamous epithelial cell |
CL:0000076 |
skos:exactMatch |
squamous epithelial cell |
squamous epithelial cell |
CL:0000076 |
skos:exactMatch |
epithelial cell |
epithelial cell |
CL:0000066 |
skos:exactMatch |
tonsil-codex-stanford |
| 244 |
Stem |
stem cell |
stem cell |
CL:0000034 |
skos:exactMatch |
stem cell |
stem cell |
CL:0000034 |
skos:exactMatch |
epithelial cell |
epithelial cell |
CL:0000066 |
skos:exactMatch |
colon-xenium-stanford |
| 245 |
Stroma |
stromal cell |
stromal cell |
CL:0000499 |
skos:exactMatch |
stromal cell |
stromal cell |
CL:0000499 |
skos:exactMatch |
mesenchymal cell |
mesenchymal cell |
CL:0008019 |
skos:exactMatch |
esophagus-codex-stanford |
| 246 |
Stroma |
stromal cell |
stromal cell |
CL:0000499 |
skos:exactMatch |
stromal cell |
stromal cell |
CL:0000499 |
skos:exactMatch |
mesenchymal cell |
mesenchymal cell |
CL:0008019 |
skos:exactMatch |
intestine-codex-stanford |
| 247 |
Stroma |
stromal cell |
stromal cell |
CL:0000499 |
skos:exactMatch |
stromal cell |
stromal cell |
CL:0000499 |
skos:exactMatch |
mesenchymal cell |
mesenchymal cell |
CL:0008019 |
skos:exactMatch |
tonsil-codex-stanford |
| 248 |
PDPN |
stromal cell:podoplanin+ |
stromal cell:podoplanin-positive |
CL:0000499 |
skos:narrowMatch |
stromal cell |
stromal cell |
CL:0000499 |
skos:exactMatch |
mesenchymal cell |
mesenchymal cell |
CL:0008019 |
skos:exactMatch |
esophagus-codex-stanford |
| 249 |
PDPN |
stromal cell:podoplanin+ |
stromal cell:podoplanin-positive |
CL:0000499 |
skos:narrowMatch |
stromal cell |
stromal cell |
CL:0000499 |
skos:exactMatch |
mesenchymal cell |
mesenchymal cell |
CL:0008019 |
skos:exactMatch |
tonsil-codex-stanford |
| 250 |
T |
t cell |
T cell |
CL:0000084 |
skos:exactMatch |
t cell |
T cell |
CL:0000084 |
skos:exactMatch |
immune cell |
leukocyte |
CL:0000738 |
skos:exactMatch |
esophagus-codex-stanford |
| 251 |
gd T Cells |
t cell |
T cell |
CL:0000084 |
skos:exactMatch |
t cell |
T cell |
CL:0000084 |
skos:exactMatch |
immune cell |
leukocyte |
CL:0000738 |
skos:exactMatch |
oralcavity-codex-czi |
| 252 |
T |
t cell |
T cell |
CL:0000084 |
skos:exactMatch |
t cell |
T cell |
CL:0000084 |
skos:exactMatch |
immune cell |
leukocyte |
CL:0000738 |
skos:exactMatch |
tonsil-codex-stanford |
| 253 |
CD8 T |
t cell |
T cell |
CL:0000084 |
skos:exactMatch |
t cell |
T cell |
CL:0000084 |
skos:exactMatch |
immune cell |
leukocyte |
CL:0000738 |
skos:exactMatch |
skin-confocal-sorgerlab |
| 254 |
CD8+_T_cell_CD_4+_T_cell |
t cell |
T cell |
CL:0000084 |
skos:exactMatch |
t cell |
T cell |
CL:0000084 |
skos:exactMatch |
immune cell |
leukocyte |
CL:0000738 |
skos:exactMatch |
lung-codex-urmc |
| 255 |
CD4 T Cells |
t cell:cd4+ |
CD4-positive, alpha-beta T cell |
CL:0000624 |
skos:exactMatch |
t cell |
T cell |
CL:0000084 |
skos:exactMatch |
immune cell |
leukocyte |
CL:0000738 |
skos:exactMatch |
oralcavity-codex-czi |
| 256 |
CD4 T |
t cell:cd4+ |
CD4-positive, alpha-beta T cell |
CL:0000624 |
skos:exactMatch |
t cell |
T cell |
CL:0000084 |
skos:exactMatch |
immune cell |
leukocyte |
CL:0000738 |
skos:exactMatch |
skin-confocal-sorgerlab |
| 257 |
CD4+ T cell |
t cell:cd4+ |
CD4-positive, alpha-beta T cell |
CL:0000624 |
skos:exactMatch |
t cell |
T cell |
CL:0000084 |
skos:exactMatch |
immune cell |
leukocyte |
CL:0000738 |
skos:exactMatch |
intestine-codex-stanford |
| 258 |
Th |
t cell:cd4+ |
CD4-positive, alpha-beta T cell |
CL:0000624 |
skos:exactMatch |
t cell |
T cell |
CL:0000084 |
skos:exactMatch |
immune cell |
leukocyte |
CL:0000738 |
skos:exactMatch |
oralcavity-codex-czi |
| 259 |
T helper |
t cell:cd4+ |
CD4-positive, alpha-beta T cell |
CL:0000624 |
skos:exactMatch |
t cell |
T cell |
CL:0000084 |
skos:exactMatch |
immune cell |
leukocyte |
CL:0000738 |
skos:exactMatch |
colon-cycif-sorgerlab |
| 260 |
T-Helper |
t cell:cd4+ |
CD4-positive, alpha-beta T cell |
CL:0000624 |
skos:exactMatch |
t cell |
T cell |
CL:0000084 |
skos:exactMatch |
immune cell |
leukocyte |
CL:0000738 |
skos:exactMatch |
skin-celldive-ge |
| 261 |
CD4+ T-Cell |
t cell:cd4+ alpha-beta |
CD4-positive, alpha-beta T cell |
CL:0000624 |
skos:exactMatch |
t cell |
T cell |
CL:0000084 |
skos:exactMatch |
immune cell |
leukocyte |
CL:0000738 |
skos:exactMatch |
bonemarrow-codex-chop |
| 262 |
CD4+ |
t cell:cd4+ alpha-beta |
CD4-positive, alpha-beta T cell |
CL:0000624 |
skos:exactMatch |
t cell |
T cell |
CL:0000084 |
skos:exactMatch |
immune cell |
leukocyte |
CL:0000738 |
skos:exactMatch |
colon-xenium-stanford |
| 263 |
CD4+_T_cell_2 |
t cell:cd4+ alpha-beta |
CD4-positive, alpha-beta T cell |
CL:0000624 |
skos:exactMatch |
t cell |
T cell |
CL:0000084 |
skos:exactMatch |
immune cell |
leukocyte |
CL:0000738 |
skos:exactMatch |
lung-codex-urmc |
| 264 |
CD4+_T_cell_3 |
t cell:cd4+ alpha-beta |
CD4-positive, alpha-beta T cell |
CL:0000624 |
skos:exactMatch |
t cell |
T cell |
CL:0000084 |
skos:exactMatch |
immune cell |
leukocyte |
CL:0000738 |
skos:exactMatch |
lung-codex-urmc |
| 265 |
CD4_+_Tcell/macrophage |
t cell:cd4+ alpha-beta |
lung resident memory CD4-positive, alpha-beta ... |
CL:4033038 |
skos:exactMatch |
t cell |
T cell |
CL:0000084 |
skos:exactMatch |
immune cell |
leukocyte |
CL:0000738 |
skos:exactMatch |
lung-codex-urmc |
| 266 |
CD4+_T_cell_1 |
t cell:cd4+ alpha-beta |
CD4-positive, alpha-beta T cell |
CL:0000624 |
skos:exactMatch |
t cell |
T cell |
CL:0000084 |
skos:exactMatch |
immune cell |
leukocyte |
CL:0000738 |
skos:exactMatch |
lung-codex-urmc |
| 267 |
CD4+ T cell |
t cell:cd4+ alpha-beta |
CD4-positive, alpha-beta T cell |
CL:0000624 |
skos:exactMatch |
t cell |
T cell |
CL:0000084 |
skos:exactMatch |
immune cell |
leukocyte |
CL:0000738 |
skos:exactMatch |
lung-codex-urmc |
| 268 |
CD4T |
t cell:cd4+ alpha-beta |
CD4-positive, alpha-beta T cell |
CL:0000624 |
skos:exactMatch |
t cell |
T cell |
CL:0000084 |
skos:exactMatch |
immune cell |
leukocyte |
CL:0000738 |
skos:exactMatch |
maternalfetalinterface-mibitof-stanford |
| 269 |
T_CD4+ |
t cell:cd4+ alpha-beta effector |
effector CD4-positive, alpha-beta T cell |
CL:0001044 |
skos:exactMatch |
t cell |
T cell |
CL:0000084 |
skos:exactMatch |
immune cell |
leukocyte |
CL:0000738 |
skos:exactMatch |
lymphnode-codex-yale |
| 270 |
CD4 Memory T cells |
t cell:cd4+ alpha-beta memory |
CD4-positive, alpha-beta memory T cell |
CL:0000897 |
skos:exactMatch |
t cell |
T cell |
CL:0000084 |
skos:exactMatch |
immune cell |
leukocyte |
CL:0000738 |
skos:exactMatch |
spleen-codex-ufl |
| 271 |
T_CD4+_naive |
t cell:cd4+ alpha-beta naive thymus-derived |
naive thymus-derived CD4-positive, alpha-beta ... |
CL:0000895 |
skos:exactMatch |
t cell |
T cell |
CL:0000084 |
skos:exactMatch |
immune cell |
leukocyte |
CL:0000738 |
skos:exactMatch |
lymphnode-codex-yale |
| 272 |
PD1+ T helper |
t cell:cd4+ pdl1+ |
CD4-positive, alpha-beta T cell:pdl1-positive |
CL:0000624 |
skos:narrowMatch |
t cell |
T cell |
CL:0000084 |
skos:exactMatch |
immune cell |
leukocyte |
CL:0000738 |
skos:exactMatch |
colon-cycif-sorgerlab |
| 273 |
T-Killer |
t cell:cd8+ |
CD8-positive, alpha-beta T cell |
CL:0000625 |
skos:exactMatch |
t cell |
T cell |
CL:0000084 |
skos:exactMatch |
immune cell |
leukocyte |
CL:0000738 |
skos:exactMatch |
skin-celldive-ge |
| 274 |
CD8+ T cell_2 |
t cell:cd8+ |
lung resident memory CD8-positive, alpha-beta ... |
CL:4033039 |
skos:exactMatch |
t cell |
T cell |
CL:0000084 |
skos:exactMatch |
immune cell |
leukocyte |
CL:0000738 |
skos:exactMatch |
lung-codex-urmc |
| 275 |
CD8 T Cells |
t cell:cd8+ |
CD8-positive, alpha-beta T cell |
CL:0000625 |
skos:exactMatch |
t cell |
T cell |
CL:0000084 |
skos:exactMatch |
immune cell |
leukocyte |
CL:0000738 |
skos:exactMatch |
oralcavity-codex-czi |
| 276 |
CD8+_T_cell_1 |
t cell:cd8+ |
lung resident memory CD8-positive, alpha-beta ... |
CL:4033039 |
skos:exactMatch |
t cell |
T cell |
CL:0000084 |
skos:exactMatch |
immune cell |
leukocyte |
CL:0000738 |
skos:exactMatch |
lung-codex-urmc |
| 277 |
T_CD8+_cytotoxic |
t cell:cd8+ |
CD8-positive, alpha-beta T cell |
CL:0000625 |
skos:exactMatch |
t cell |
T cell |
CL:0000084 |
skos:exactMatch |
immune cell |
leukocyte |
CL:0000738 |
skos:exactMatch |
lymphnode-codex-yale |
| 278 |
T_CD8+_naive |
t cell:cd8+ |
CD8-positive, alpha-beta T cell |
CL:0000625 |
skos:exactMatch |
t cell |
T cell |
CL:0000084 |
skos:exactMatch |
immune cell |
leukocyte |
CL:0000738 |
skos:exactMatch |
lymphnode-codex-yale |
| 279 |
Tc |
t cell:cd8+ |
CD8-positive, alpha-beta T cell |
CL:0000625 |
skos:exactMatch |
t cell |
T cell |
CL:0000084 |
skos:exactMatch |
immune cell |
leukocyte |
CL:0000738 |
skos:exactMatch |
oralcavity-codex-czi |
| 280 |
CD8 + T cell_1 |
t cell:cd8+ |
lung resident memory CD8-positive, alpha-beta ... |
CL:4033039 |
skos:exactMatch |
t cell |
T cell |
CL:0000084 |
skos:exactMatch |
immune cell |
leukocyte |
CL:0000738 |
skos:exactMatch |
lung-codex-urmc |
| 281 |
CD8+_T_cell_3 |
t cell:cd8+ |
lung resident memory CD8-positive, alpha-beta ... |
CL:4033039 |
skos:exactMatch |
t cell |
T cell |
CL:0000084 |
skos:exactMatch |
immune cell |
leukocyte |
CL:0000738 |
skos:exactMatch |
lung-codex-urmc |
| 282 |
Tc cell |
t cell:cd8+ |
CD8-positive, alpha-beta T cell |
CL:0000625 |
skos:exactMatch |
t cell |
T cell |
CL:0000084 |
skos:exactMatch |
immune cell |
leukocyte |
CL:0000738 |
skos:exactMatch |
colon-cycif-sorgerlab |
| 283 |
CD8+_T_cell_2 |
t cell:cd8+ |
lung resident memory CD8-positive, alpha-beta ... |
CL:4033039 |
skos:exactMatch |
t cell |
T cell |
CL:0000084 |
skos:exactMatch |
immune cell |
leukocyte |
CL:0000738 |
skos:exactMatch |
lung-codex-urmc |
| 284 |
CD8+ T |
t cell:cd8+ alpha-beta |
CD8-positive, alpha-beta T cell |
CL:0000625 |
skos:exactMatch |
t cell |
T cell |
CL:0000084 |
skos:exactMatch |
immune cell |
leukocyte |
CL:0000738 |
skos:exactMatch |
intestine-codex-stanford |
| 285 |
CD8T |
t cell:cd8+ alpha-beta |
CD8-positive, alpha-beta T cell |
CL:0000625 |
skos:exactMatch |
t cell |
T cell |
CL:0000084 |
skos:exactMatch |
immune cell |
leukocyte |
CL:0000738 |
skos:exactMatch |
maternalfetalinterface-mibitof-stanford |
| 286 |
CD8+ |
t cell:cd8+ alpha-beta effector memory |
effector memory CD8-positive, alpha-beta T cell |
CL:0000913 |
skos:exactMatch |
t cell |
T cell |
CL:0000084 |
skos:exactMatch |
immune cell |
leukocyte |
CL:0000738 |
skos:exactMatch |
colon-xenium-stanford |
| 287 |
CD8 Memory T cells |
t cell:cd8+ alpha-beta memory |
CD8-positive, alpha-beta memory T cell |
CL:0000909 |
skos:exactMatch |
t cell |
T cell |
CL:0000084 |
skos:exactMatch |
immune cell |
leukocyte |
CL:0000738 |
skos:exactMatch |
spleen-codex-ufl |
| 288 |
CD8+ T-Cell |
t cell:cd8+ alpha-beta regulatory |
CD8-positive, alpha-beta regulatory T cell |
CL:0000795 |
skos:exactMatch |
t cell |
T cell |
CL:0000084 |
skos:exactMatch |
immune cell |
leukocyte |
CL:0000738 |
skos:exactMatch |
bonemarrow-codex-chop |
| 289 |
T_CD8+_CD161+ |
t cell:cd8+ cd161+ |
CD8-positive, alpha-beta T cell:CD161-positive |
CL:0000625 |
skos:narrowMatch |
t cell |
T cell |
CL:0000084 |
skos:exactMatch |
immune cell |
leukocyte |
CL:0000738 |
skos:exactMatch |
lymphnode-codex-yale |
| 290 |
PD1+ Tc |
t cell:cd8+ pdl1+ |
CD8-positive, alpha-beta T cell:pdl1-positive |
CL:0000625 |
skos:narrowMatch |
t cell |
T cell |
CL:0000084 |
skos:exactMatch |
immune cell |
leukocyte |
CL:0000738 |
skos:exactMatch |
colon-cycif-sorgerlab |
| 291 |
T_CD4+_TfH_GC |
t cell:follicular helper |
T follicular helper cell |
CL:0002038 |
skos:exactMatch |
t cell |
T cell |
CL:0000084 |
skos:exactMatch |
immune cell |
leukocyte |
CL:0000738 |
skos:exactMatch |
lymphnode-codex-yale |
| 292 |
T_CD4+_TfH |
t cell:follicular helper |
T follicular helper cell |
CL:0002038 |
skos:exactMatch |
t cell |
T cell |
CL:0000084 |
skos:exactMatch |
immune cell |
leukocyte |
CL:0000738 |
skos:exactMatch |
lymphnode-codex-yale |
| 293 |
NKT |
t cell:mature natural killer |
mature NK T cell |
CL:0000814 |
skos:exactMatch |
t cell |
T cell |
CL:0000084 |
skos:exactMatch |
immune cell |
leukocyte |
CL:0000738 |
skos:exactMatch |
lymphnode-codex-yale |
| 294 |
NKT |
t cell:mature natural killer |
mature NK T cell |
CL:0000814 |
skos:exactMatch |
t cell |
T cell |
CL:0000084 |
skos:exactMatch |
immune cell |
leukocyte |
CL:0000738 |
skos:exactMatch |
maternalfetalinterface-mibitof-stanford |
| 295 |
Tissue T |
t cell:memory |
memory T cell |
CL:0000813 |
skos:exactMatch |
t cell |
T cell |
CL:0000084 |
skos:exactMatch |
immune cell |
leukocyte |
CL:0000738 |
skos:exactMatch |
skin-confocal-sorgerlab |
| 296 |
Naive T |
t cell:naive |
naive T cell |
CL:0000898 |
skos:exactMatch |
t cell |
T cell |
CL:0000084 |
skos:exactMatch |
immune cell |
leukocyte |
CL:0000738 |
skos:exactMatch |
colon-xenium-stanford |
| 297 |
T reg |
t cell:regulatory |
regulatory T cell |
CL:0000815 |
skos:exactMatch |
t cell |
T cell |
CL:0000084 |
skos:exactMatch |
immune cell |
leukocyte |
CL:0000738 |
skos:exactMatch |
skin-confocal-sorgerlab |
| 298 |
Treg |
t cell:regulatory |
regulatory T cell |
CL:0000815 |
skos:exactMatch |
t cell |
T cell |
CL:0000084 |
skos:exactMatch |
immune cell |
leukocyte |
CL:0000738 |
skos:exactMatch |
oralcavity-codex-czi |
| 299 |
Treg |
t cell:regulatory |
regulatory T cell |
CL:0000815 |
skos:exactMatch |
t cell |
T cell |
CL:0000084 |
skos:exactMatch |
immune cell |
leukocyte |
CL:0000738 |
skos:exactMatch |
maternalfetalinterface-mibitof-stanford |
| 300 |
Treg |
t cell:regulatory |
regulatory T cell |
CL:0000815 |
skos:exactMatch |
t cell |
T cell |
CL:0000084 |
skos:exactMatch |
immune cell |
leukocyte |
CL:0000738 |
skos:exactMatch |
colon-cycif-sorgerlab |
| 301 |
T_Treg |
t cell:regulatory |
regulatory T cell |
CL:0000815 |
skos:exactMatch |
t cell |
T cell |
CL:0000084 |
skos:exactMatch |
immune cell |
leukocyte |
CL:0000738 |
skos:exactMatch |
lymphnode-codex-yale |
| 302 |
T-Reg |
t cell:regulatory |
regulatory T cell |
CL:0000815 |
skos:exactMatch |
t cell |
T cell |
CL:0000084 |
skos:exactMatch |
immune cell |
leukocyte |
CL:0000738 |
skos:exactMatch |
skin-celldive-ge |
| 303 |
Tregs |
t cell:regulatory |
regulatory T cell |
CL:0000815 |
skos:exactMatch |
t cell |
T cell |
CL:0000084 |
skos:exactMatch |
immune cell |
leukocyte |
CL:0000738 |
skos:exactMatch |
colon-xenium-stanford |
| 304 |
T_TfR |
t cell:regulatory |
regulatory T cell |
CL:0000815 |
skos:exactMatch |
t cell |
T cell |
CL:0000084 |
skos:exactMatch |
immune cell |
leukocyte |
CL:0000738 |
skos:exactMatch |
lymphnode-codex-yale |
| 305 |
T_TIM3+ |
t cell:tim3+ |
T cell:tim3-positive |
CL:0000084 |
skos:narrowMatch |
t cell |
T cell |
CL:0000084 |
skos:exactMatch |
immune cell |
leukocyte |
CL:0000738 |
skos:exactMatch |
lymphnode-codex-yale |
| 306 |
DP |
thymocyte:cd4-intermediate cd8+ double-positive |
CD4-intermediate, CD8-positive double-positive... |
CL:0002430 |
skos:exactMatch |
t cell |
T cell |
CL:0000084 |
skos:exactMatch |
immune cell |
leukocyte |
CL:0000738 |
skos:exactMatch |
oralcavity-codex-czi |
| 307 |
TA2 |
transit amplifying cell |
transit amplifying cell |
CL:0009010 |
skos:exactMatch |
enterocyte |
enterocyte |
CL:0000584 |
skos:exactMatch |
epithelial cell |
epithelial cell |
CL:0000066 |
skos:exactMatch |
colon-xenium-stanford |
| 308 |
TA |
transit amplifying cell |
transit amplifying cell |
CL:0009010 |
skos:exactMatch |
enterocyte |
enterocyte |
CL:0000584 |
skos:exactMatch |
epithelial cell |
epithelial cell |
CL:0000066 |
skos:exactMatch |
intestine-codex-stanford |
| 309 |
TA1 |
transit amplifying cell |
transit amplifying cell |
CL:0009010 |
skos:exactMatch |
enterocyte |
enterocyte |
CL:0000584 |
skos:exactMatch |
epithelial cell |
epithelial cell |
CL:0000066 |
skos:exactMatch |
colon-xenium-stanford |
| 310 |
CyclingTA |
transit amplifying cell:proliferating |
transit amplifying cell:proliferating |
CL:0009010 |
skos:narrowMatch |
transit amplifying cell |
transit amplifying cell |
CL:0009010 |
skos:exactMatch |
immune cell |
leukocyte |
CL:0000738 |
skos:exactMatch |
colon-xenium-stanford |
| 311 |
Cycling TA |
transit amplifying cell:proliferating |
transit amplifying cell:proliferating |
CL:0009010 |
skos:narrowMatch |
enterocyte |
enterocyte |
CL:0000584 |
skos:exactMatch |
epithelial cell |
epithelial cell |
CL:0000066 |
skos:exactMatch |
intestine-codex-stanford |
| 312 |
EVT2 |
trophoblast:hla-g minus, cd57- ck7 low extravi... |
extravillous trophoblast:hla-g-negative cd57-n... |
CL:0008036 |
skos:narrowMatch |
trophoblast |
trophoblast cell |
CL:0000351 |
skos:exactMatch |
epithelial cell |
epithelial cell |
CL:0000066 |
skos:exactMatch |
maternalfetalinterface-mibitof-stanford |
| 313 |
EVT1c |
trophoblast:hla-g+ cd56+ extravillous |
extravillous trophoblast:hla-g-positive cd56-p... |
CL:0008036 |
skos:narrowMatch |
trophoblast |
trophoblast cell |
CL:0000351 |
skos:exactMatch |
epithelial cell |
epithelial cell |
CL:0000066 |
skos:exactMatch |
maternalfetalinterface-mibitof-stanford |
| 314 |
EVT1a |
trophoblast:hla-g+ ck7+ extravillous |
extravillous trophoblast:hla-g-positive ck7-po... |
CL:0008036 |
skos:narrowMatch |
trophoblast |
trophoblast cell |
CL:0000351 |
skos:exactMatch |
epithelial cell |
epithelial cell |
CL:0000066 |
skos:exactMatch |
maternalfetalinterface-mibitof-stanford |
| 315 |
EVT1b |
trophoblast:hla-g+ ck7- extravillous |
extravillous trophoblast:hla-g-positive ck7-ne... |
CL:0008036 |
skos:narrowMatch |
trophoblast |
trophoblast cell |
CL:0000351 |
skos:exactMatch |
epithelial cell |
epithelial cell |
CL:0000066 |
skos:exactMatch |
maternalfetalinterface-mibitof-stanford |
| 316 |
Tuft |
tuft cell:intestinal |
intestinal tuft cell |
CL:0019032 |
skos:exactMatch |
tuft cell |
brush cell |
CL:0002204 |
skos:exactMatch |
epithelial cell |
epithelial cell |
CL:0000066 |
skos:exactMatch |
colon-xenium-stanford |
| 317 |
Tumor |
tumor cell |
neoplastic cell |
CL:0001063 |
skos:exactMatch |
abnormal cell |
abnormal cell |
CL:0001061 |
skos:exactMatch |
unknown cell |
cell:unknown |
CL:0000000 |
skos:narrowMatch |
skin-confocal-sorgerlab |
| 318 |
indistinct |
unknown cell |
cell:unknown |
CL:0000000 |
skos:narrowMatch |
unknown cell |
cell:unknown |
CL:0000000 |
skos:narrowMatch |
unknown cell |
cell:unknown |
CL:0000000 |
skos:narrowMatch |
spleen-codex-ufl |
| 319 |
Others |
unknown cell |
cell:unknown |
CL:0000000 |
skos:narrowMatch |
unknown cell |
cell:unknown |
CL:0000000 |
skos:narrowMatch |
unknown cell |
cell:unknown |
CL:0000000 |
skos:narrowMatch |
oralcavity-codex-czi |
| 320 |
UNK_3 |
unknown cell |
cell:unknown |
CL:0000000 |
skos:narrowMatch |
unknown cell |
cell:unknown |
CL:0000000 |
skos:narrowMatch |
unknown cell |
cell:unknown |
CL:0000000 |
skos:narrowMatch |
lung-codex-urmc |
| 321 |
UNK_2 |
unknown cell |
cell:unknown |
CL:0000000 |
skos:narrowMatch |
unknown cell |
cell:unknown |
CL:0000000 |
skos:narrowMatch |
unknown cell |
cell:unknown |
CL:0000000 |
skos:narrowMatch |
lung-codex-urmc |
| 322 |
UNK_5_ambiguous |
unknown cell |
cell:unknown |
CL:0000000 |
skos:narrowMatch |
unknown cell |
cell:unknown |
CL:0000000 |
skos:narrowMatch |
unknown cell |
cell:unknown |
CL:0000000 |
skos:narrowMatch |
lung-codex-urmc |
| 323 |
UNK_1_APC |
unknown cell |
cell:unknown |
CL:0000000 |
skos:narrowMatch |
unknown cell |
cell:unknown |
CL:0000000 |
skos:narrowMatch |
unknown cell |
cell:unknown |
CL:0000000 |
skos:narrowMatch |
lung-codex-urmc |
| 324 |
UNK_3_(col1a1-driven_cluster) |
unknown cell |
cell:unknown |
CL:0000000 |
skos:narrowMatch |
unknown cell |
cell:unknown |
CL:0000000 |
skos:narrowMatch |
unknown cell |
cell:unknown |
CL:0000000 |
skos:narrowMatch |
lung-codex-urmc |
| 325 |
UNK_4_(col1a1_driven_cluster) |
unknown cell |
cell:unknown |
CL:0000000 |
skos:narrowMatch |
unknown cell |
cell:unknown |
CL:0000000 |
skos:narrowMatch |
unknown cell |
cell:unknown |
CL:0000000 |
skos:narrowMatch |
lung-codex-urmc |
| 326 |
Unknown |
unknown cell |
cell:unknown |
CL:0000000 |
skos:narrowMatch |
unknown cell |
cell:unknown |
CL:0000000 |
skos:narrowMatch |
unknown cell |
cell:unknown |
CL:0000000 |
skos:narrowMatch |
colon-xenium-stanford |
| 327 |
other |
unknown cell |
cell:unknown |
CL:0000000 |
skos:narrowMatch |
unknown cell |
cell:unknown |
CL:0000000 |
skos:narrowMatch |
unknown cell |
cell:unknown |
CL:0000000 |
skos:narrowMatch |
maternalfetalinterface-mibitof-stanford |
| 328 |
UNK_1 |
unknown cell |
cell:unknown |
CL:0000000 |
skos:narrowMatch |
unknown cell |
cell:unknown |
CL:0000000 |
skos:narrowMatch |
unknown cell |
cell:unknown |
CL:0000000 |
skos:narrowMatch |
lung-codex-urmc |
| 329 |
Unknown_lowcount |
unknown cell |
cell:unknown |
CL:0000000 |
skos:narrowMatch |
unknown cell |
cell:unknown |
CL:0000000 |
skos:narrowMatch |
unknown cell |
cell:unknown |
CL:0000000 |
skos:narrowMatch |
colon-xenium-stanford |
| 330 |
unknown |
unknown cell |
cell:unknown |
CL:0000000 |
skos:narrowMatch |
unknown cell |
cell:unknown |
CL:0000000 |
skos:narrowMatch |
unknown cell |
cell:unknown |
CL:0000000 |
skos:narrowMatch |
pancreas-geomx-ufl |
| 331 |
Artifact |
unknown cell |
cell:unknown |
CL:0000000 |
skos:narrowMatch |
unknown cell |
cell:unknown |
CL:0000000 |
skos:narrowMatch |
unknown cell |
cell:unknown |
CL:0000000 |
skos:narrowMatch |
bonemarrow-codex-chop |
| 332 |
Undetermined |
unknown cell |
cell:unknown |
CL:0000000 |
skos:narrowMatch |
unknown cell |
cell:unknown |
CL:0000000 |
skos:narrowMatch |
unknown cell |
cell:unknown |
CL:0000000 |
skos:narrowMatch |
bonemarrow-codex-chop |
| 333 |
Autofluorescent |
unknown cell |
cell:unknown |
CL:0000000 |
skos:narrowMatch |
unknown cell |
cell:unknown |
CL:0000000 |
skos:narrowMatch |
unknown cell |
cell:unknown |
CL:0000000 |
skos:narrowMatch |
bonemarrow-codex-chop |
| 334 |
CD44+ Undetermined |
unknown cell |
cell:unknown |
CL:0000000 |
skos:narrowMatch |
unknown cell |
cell:unknown |
CL:0000000 |
skos:narrowMatch |
unknown cell |
cell:unknown |
CL:0000000 |
skos:narrowMatch |
bonemarrow-codex-chop |
| 335 |
Other |
unknown cell |
cell:unknown |
CL:0000000 |
skos:narrowMatch |
unknown cell |
cell:unknown |
CL:0000000 |
skos:narrowMatch |
unknown cell |
cell:unknown |
CL:0000000 |
skos:narrowMatch |
colon-cycif-sorgerlab |
| 336 |
ENDO_CD8+_T_Cell |
unknown cell |
cell:unknown |
CL:0000000 |
skos:narrowMatch |
unknown cell |
cell:unknown |
CL:0000000 |
skos:narrowMatch |
unknown cell |
cell:unknown |
CL:0000000 |
skos:narrowMatch |
lung-codex-urmc |
| 337 |
Unknown |
unknown cell |
cell:unknown |
CL:0000000 |
skos:narrowMatch |
unknown cell |
cell:unknown |
CL:0000000 |
skos:narrowMatch |
unknown cell |
cell:unknown |
CL:0000000 |
skos:narrowMatch |
skin-confocal-sorgerlab |